Figure 1.
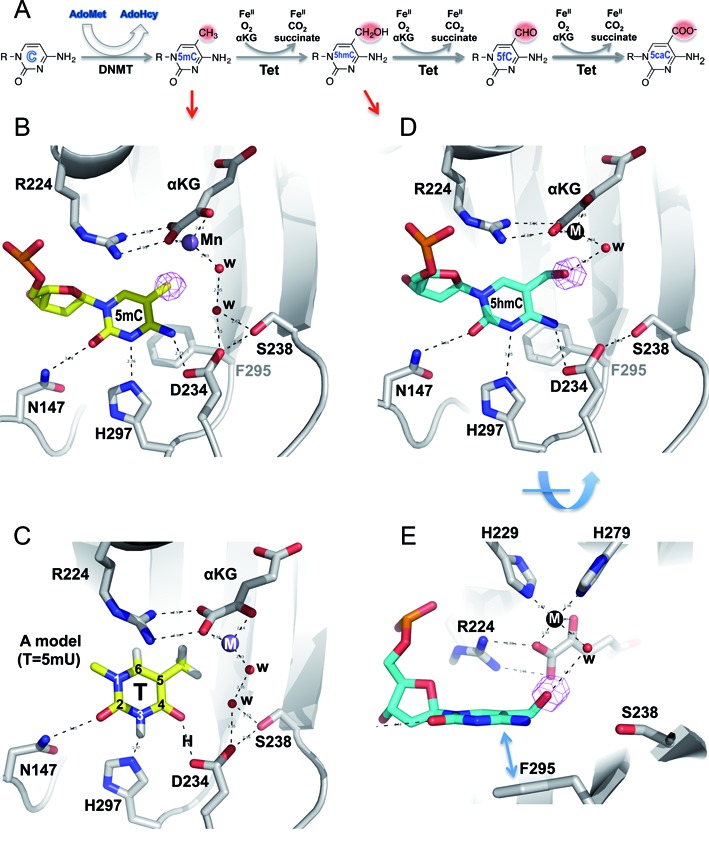
Structures of NgTet1 in complexes with 5mC and 5hmC DNA. (A) Schematic illustration of methylation and oxidation reactions. DNA methyltransferases convert a proportion of the cytosines (Cs) into 5mC in a S-adenosyl-l-methionine (AdoMet)-dependent reaction. The Tet dioxygenases then convert a fraction of 5mC to 5hmC, 5fC and 5caC in three consecutive, Fe(II)- and α-ketoglutarate-dependent oxidation reactions without releasing any formaldehyde (in contrast to demethylation of N-methylated substrates; Supplemental Figure S1). (B) The extrahelical 5mC in the active site forms planar π stacking contacts with F295 (away from the viewer in the background) and R224 (which forms an ion-pair interaction with α-ketoglutarate or αKG), as well as hydrogen bonds with three residues along the Watson–Crick polar edge. The simulated annealing omit electron density (in magenta) is shown for the methyl group of 5mC, contoured at 4.5σ above the mean. (C) A model of thymine (5mU) in the active site including hydrogen atoms. The side-chain imidazole ring of H297 could interact with the protonated N3 nitrogen. The distance between one of the carboxylate oxygen atoms of D234 and the O4 carbonyl oxygen would suggest the presence of a hydrogen bond and therefore the presence of a proton between them (labeled as H). The proton source may be the COOH group of D234, which itself might be protonated as a result of the water-mediated interaction. (D) The extrahelical 5hmC in the active site forms nearly identical interaction as that of 5mC (panel B). The simulated annealing omit electron density (in magenta) for the hydroxyl oxygen atom of 5hmC is shown, contoured at 4.5σ above the mean. (E) An orthogonal view from panel D shows the out-of-plane hydroxyl oxygen atom of 5hmC interacting with the metal-ligand water molecule.
