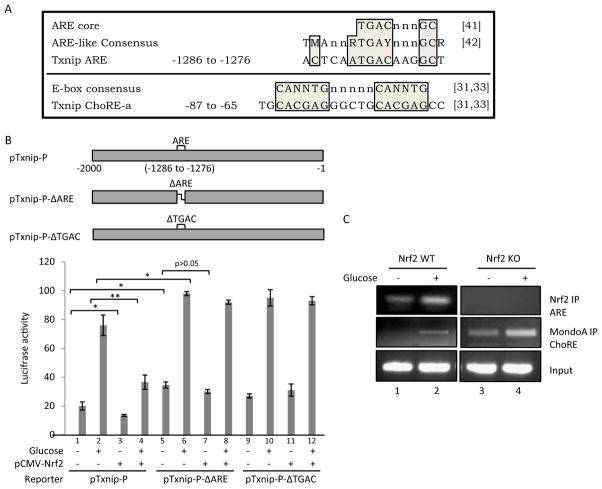
Fig. 6.
Interaction between Nrf2 and Txnip ARE. A, sequences of ARE and ChoRE-a of Txnip and alignment with ARE core and E-box consensus. A putative ARE sequence was identified in Txnip promoter (−1286 to −1276) that matches the ARE core sequence and is highly similar to the ARE-like sequence consensus. Shown also is the Txnip ChoRE-a sequence and its alignment with E-box consensus. N or n = G, C, T, or A; M = A or C; R = A or G; and Y = C or T. Positions represent base pairs upstream of the transcription start site of mouse Txnip. References are shown in brackets: [41] = Nguyen et al. (2003); [42] = Hayes et al. (2010); and [31,33] = Minn et al. (2005). B, mutational analysis of the Txnip promoter. pTxnip-P or a deletion mutant of Txnip promoter, pTxnip-P-ΔARE or pTxnip-P-ΔTGAC, was transfected into H9C2 cells with or without pCMV-Nrf2 and glucose treatment as described for Fig. 5. Luciferase activities were measured and are expressed as means and STD from three samples. Statistical significance was shown for some comparisons; statistical significance for the pTxnip-P-ΔTGAC group (lanes 9–12) (not shown) is similar to that for the pTxnip-P- ΔARE group (lanes 5–8). *, p < 0.05; **, p < 0.01. C, ChIP assay. H9C2 cells were treated with glucose. ChIP analyses were performed to measure binding of Nrf2 to the putative Txnip ARE and of MondoA to ChoRE-a.