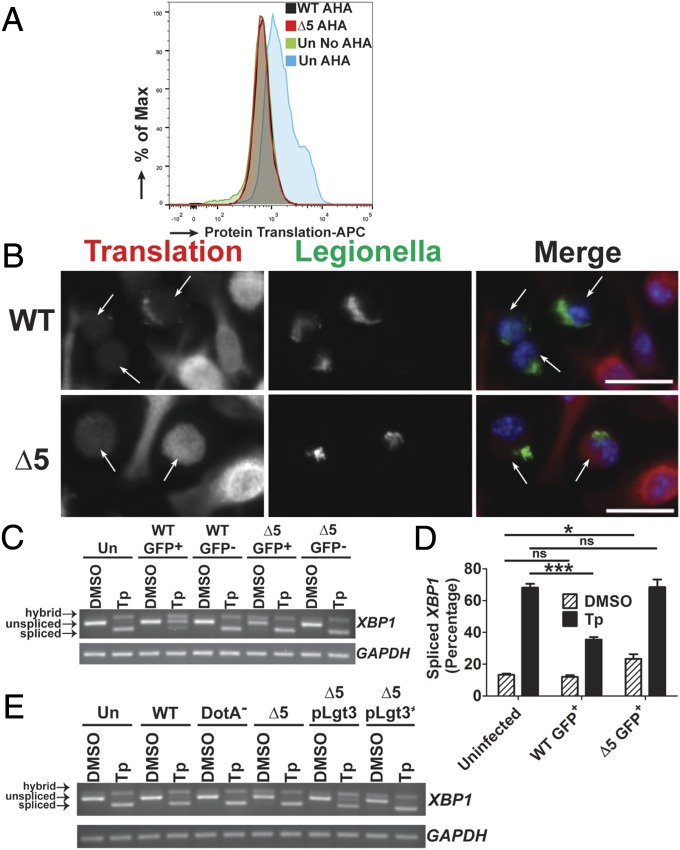
Fig. 5.
The mechanism of translation inhibition is critical for blocking XBP1 splicing. (A) The ∆5 strain inhibits protein synthesis at late time points during infection. U937 cells were challenged with GFP-L. pneumophila strains at an MOI of 1 for 10 h, then incubated with the methionine analog AHA (100 µM) for 1 h. Fixed cells harboring GFP-expressing WT or ∆5 were analyzed for levels of translation by detection of AHA incorporation into nascent peptides using DyLight 650-phosphine and flow cytometry (Materials and Methods). (B) Low levels of translation are observed at late time points during challenge with the ∆5 strain when analyzed by puromycin incorporation into translating ribosomes. A/J BMDMs were challenged with bacteria at an MOI of 0.5 for 10 h then treated with puromycin (1 µg/mL) for another 1 h. Cells were fixed and permeablized, then stained with antibodies specific to L. pneumophila or puromycin, to measure translation (Materials and Methods). (Scale bar: 10 µm.) (C) At late time points, the ∆5 strain induces XBP1 splicing and is unable to inhibit chemically induced XBP1 splicing. U937 cells were challenged with bacteria at an MOI of 1 for 9 h, then treated with either DMSO or Tp (100 nM) for 2 h. Cells were sorted by GFP (for the infected populations), and lysates from each population were used to isolate RNA (Materials and Methods). (D) Quantitation of XBP1 splicing from C. (E) Expression of Lgt3 limits chemically induced XBP1 splicing at late time points. U937 cells were challenged with each strain at an MOI of 1 for 11 h and then treated with DMSO or Tp (100 nM) at 9 h postchallenge. RNA isolated from the total cell population was analyzed for XBP1 splicing (Materials and Methods). Data are representative of three independent experiments (A–C and E) or the mean ± SEM of three independent experiments (D). Statistical analyses were performed using unpaired t test. *P < 0.05; ***P < 0.001.