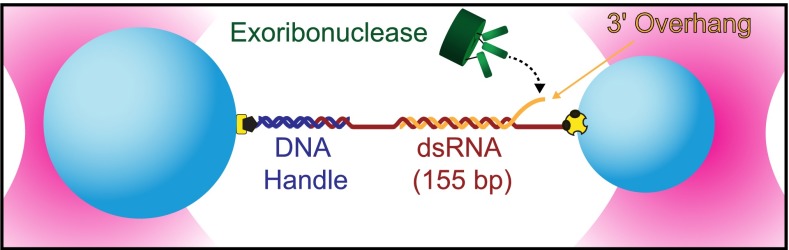
Fig. 1.
Single-molecule exoribonuclease assay (not to scale). A 155-bp segment of dsRNA is placed under tension between polystyrene beads (light blue) held in twin optical traps (pink). One strand of RNA (the substrate, orange) carries a 3′ adenine-rich terminal overhang that serves as a loading site for a processive exoribonuclease (green) that unwinds in the 3′-to-5′ direction. The complementary strand (red) is attached at its ends to the beads directly at its 5′ terminus via a biotin–avidin linkage (yellow and black) and via hybridization to a DNA handle (dark blue) attached with a digoxygenin–antibody linkage (yellow and black). As the exoribonuclease unwinds and digests the RNA substrate, the length of the duplex region is reduced, resulting in an overall increase in the tether extension. In some experiments, the RNA complementary to the enzyme substrate was replaced with DNA.