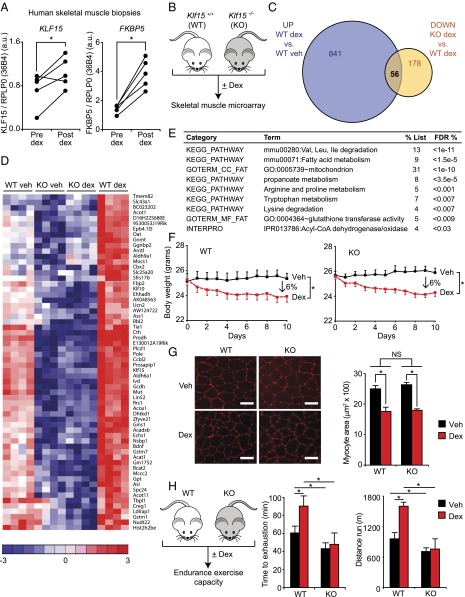
Fig. 1.
The GC–KLF15 axis dissociates ergogenic physiology from muscle atrophy. (A) KLF15 expression in skeletal muscle of healthy human subjects pre- and postdex administration. FKBP5 is a known direct GR target shown as a positive control (n = 5; *P < 0.05 for indicated comparisons). (B) Schematic for skeletal muscle (quadriceps) microarray study (n = 4 per group). (C) Venn diagram of dex-inducible and KLF15-regulated genes [2 mg/kg i.p. × 1 dose, 8 h time point; fold change > 1.5 and false discovery rate (FDR) < 0.05]. (D) Heat map of genes robustly regulated [family-wise error rate (FWER) < 0.05] by GC–KLF15 axis in mouse skeletal muscle. (E) Functional annotation [Database for Annotation, Visualization, and Integrated Discovery (DAVID)] of genes regulated by GC–KLF15 axis. (F) Body weight and (G) muscle (tibialis anterior) histology [wheat-germ agglutinin (WGA) stain] and cross-sectional area in a mouse model of chronic dex-induced atrophy (1 mg/kg·d s.c. × 10 d; n = 8; *P < 0.05 for indicated comparisons). (Scale bar, 50 µm.) (H) Schematic of dex-induced doping experiment (2 mg/kg i.p. × 1 dose) with time to exhaustion and distance run on a motorized treadmill 18 h post-dex administration (n = 7–9; *P < 0.05 for indicated comparisons). Data are shown as mean ± SEM.