Significance
We report evidence that microbes are trophically equivalent to animals. When bacteria or fungi are fed the same diets as animals, the microbes register the same trophic position as animals. This discovery reframes how microbes can be viewed within food chains and facilitates the inclusion of the microbiome in functional diversity studies. To demonstrate the broad applicability of our approach, we investigated the ancient symbioses represented by leaf-cutter ant fungus gardens, revealing four discrete trophic levels within this community and providing evidence that fungi, not ants, are the dominant herbivores of the Neotropics. Altogether, we show that microbes can be integrated with plants and animals in a food chain, thereby unifying the macro- and microbiome in studies of trophic ecology.
Keywords: compound specific, food chain, leaf-cutter ant, microbe, stable isotope
Abstract
In most ecosystems, microbes are the dominant consumers, commandeering much of the heterotrophic biomass circulating through food webs. Characterizing functional diversity within the microbiome, therefore, is critical to understanding ecosystem functioning, particularly in an era of global biodiversity loss. Using isotopic fingerprinting, we investigated the trophic positions of a broad diversity of heterotrophic organisms. Specifically, we examined the naturally occurring stable isotopes of nitrogen (15N:14N) within amino acids extracted from proteobacteria, actinomycetes, ascomycetes, and basidiomycetes, as well as from vertebrate and invertebrate macrofauna (crustaceans, fish, insects, and mammals). Here, we report that patterns of intertrophic 15N-discrimination were remarkably similar among bacteria, fungi, and animals, which permitted unambiguous measurement of consumer trophic position, independent of phylogeny or ecosystem type. The observed similarities among bacterial, fungal, and animal consumers suggest that within a trophic hierarchy, microbiota are equivalent to, and can be interdigitated with, macrobiota. To further test the universality of this finding, we examined Neotropical fungus gardens, communities in which bacteria, fungi, and animals are entwined in an ancient, quadripartite symbiosis. We reveal that this symbiosis is a discrete four-level food chain, wherein bacteria function as the apex carnivores, animals and fungi are meso-consumers, and the sole herbivores are fungi. Together, our findings demonstrate that bacteria, fungi, and animals can be integrated within a food chain, effectively uniting the macro- and microbiome in food web ecology and facilitating greater inclusion of the microbiome in studies of functional diversity.
The trophic functions of species sculpt the unique characteristics of the communities in which they are embedded (1, 2). It is not surprising, then, that sustained biodiversity losses (3, 4) are fueling the “trophic downgrading” of Earth (5) and altering the functioning of all major ecosystem types (6). The most abundant, ubiquitous organisms in most ecosystems—the microbiota—are also likely being impacted, and these organisms are often the least understood, particularly in terms of functional diversity (7, 8). If we are to better understand the impacts of biodiversity loss on ecosystem functioning, we will need to have a means to more comprehensively measure the trophic niches of both the macro- and microbiome. Further, such organisms must be examined while integrated within their respective communities (1). Assessing the trophic niches of microbial and animal species has too often been a theoretical endeavor (9), with macrofauna generally associated with plant-based food webs (“green food webs”) and the microfauna relegated to the sphere of decomposition (“brown food webs”) (9–11). It has proven difficult to merge these two spheres using a shared trophic metric, not only because of the difficulties in identifying microbial diversity (8), but also because of methodological obstacles in measuring the trophic position of a microbe. Consequently, trophic ecology has tended to focus on green food webs, often leaving the trophic hierarchies of brown webs less resolved.
Uniting green and brown food webs is critically important to studies of biodiversity and ecosystem functioning because in most ecosystems, microbes are the dominant heterotrophs (3, 7, 12). These organisms commandeer most of the heterotrophic biomass circulating through the food web (13). Indeed, in terrestrial systems, the vast majority of primary production is not captured by herbivores; rather, it falls to the ground and is consumed by microbes and small invertebrate detritivores (7, 12, 14). Higher-order carnivores consume the detritivores, conjoining the upward flow of detritivore and herbivore biomass (9, 11, 15, 16), but if the trophic positions in the basal layers of the food web cannot be accurately measured, the entire food web rests on a poorly known, tenuous platform (9).
Here, we investigate whether the trophic positions of microorganisms can be measured empirically, testing whether microbes are trophically equivalent to the macrobiotic consumers in a food-chain. Using amino acid stable isotope fingerprinting (17–20), we measure with high accuracy the trophic positions (TPs) of cultured and free-roaming organisms. In past work, this method has produced accurate TP estimates of zooplankton (18, 21), fish (17), gastropods (17), amphibians (22), and insects (19, 20), ranging from simple herbivores to higher-order carnivores. Although the utility of isotopic fingerprinting has been demonstrated in both aquatic (17, 21, 23, 24) and terrestrial ecosystems (19, 20), there remains an unbridged knowledge gap between the macro- and microbiome.
Recent studies have documented evidence of feeding guilds among the animals of brown food webs (11, 15, 25), and this finding sets up questions as to the trophic positions of the resident microbiota. For example, when microbes feed on a resource, do they register trophically just as animals do? More specifically, what is the trophic position of a fungus consuming a leaf, or an arthropod consuming that fungus? We address these questions, testing the hypothesis that fungi and bacteria are trophically equivalent to animals. We do so by measuring the degree of intertrophic 15N-discrimination within amino acids extracted from consumer taxa, focusing on two key compounds: glutamic acid and phenylalanine (17, 19, 20, 26). For microbial organisms to be trophically equivalent to animals, their respective patterns of intertrophic 15N-discrimination in glutamic acid and phenylalanine must mirror those of animals. In multiple controlled-feeding studies involving a diversity of vertebrate and invertebrate consumers, net 15N-discrimination between glutamic acid (glu) and phenylalanine (phe), commonly referred to as the trophic discrimination factor (TDFglu-phe) (27), has tended to be centered near +7.64 ± 0.60‰ (17, 20, 21). Although centered at 7.64, variability in the TDFglu-phe can generate “noise” around the trophic position (TPglu-phe) estimate for any given consumer (Eq. 1). Variability in the TDFglu-phe among consumer groups, therefore, must be assessed using known, homogeneous diets. Ultimately, the degree to which the TDFglu-phe is consistent among microbial and animal consumers will dictate whether macro- and microbiota can be considered trophically equivalent.
To assess the universality of our approach, we examine the trophic identities of the organisms in leaf-cutter ant fungus gardens. Fungus gardens represent discrete community modules in which animals (ants), fungi, and bacteria have coevolved within an ancient symbiosis (28, 29) (Fig. 1). Leaf-cutter ants (Acromyrmex or Atta) are prodigious harvesters of leaf material and have been referred to as the dominant herbivores of the Neotropics (30). The success of leaf-cutter ants derives largely from their mutualism with the fungus, Leucoagaricus gongylophorous, which is cultivated within the fungus gardens as the ants’ sole food source (31). This mutualism is frequently parasitized, however, by another fungus (Escovopsis) that can invade and overwhelm the colony (28). Leaf-cutter ants (specifically, Acromyrmex) have evolved a defense against Escovopsis via a second mutualism with a bacterium (Pseudonocardia) that selectively suppresses the growth of the invading fungus (28, 29). The fungus garden, therefore, represents a complex, quadripartite symbiosis (ant-fungus-fungus-bacterium), with major impacts on tropical food web ecology (29, 30). Our findings uncover distinct trophic identities within this community, reframing how microbes may be viewed within a food web, and demonstrating the ecological portability of our approach.
Fig. 1.
Denizens of a leaf-cutter ant fungus garden. (A) Forager ant cutting out a leaf fragment. (B) Incorporation of leaf material into the fungus garden. (C) Mycelia and fruiting bodies of Leucoagaricus, the fungus cultivated by the ants. (D) The bacterium, Pseudonocardia (white, powder-like substance on the ant dorsum), growing within specialized structures on the ant cuticle. The five taxa in our fungus gardens: oak (Quercus macrocarpa), cultivated fungi (Basidiomycota: Leucoagaricus gongylophorus), parasitic fungi (Ascomycota: Escovopsis), leaf-cutter ants (Acromyrmex echinatior), and the filamentous bacterium (Actinobacteria: Pseudonocardia) grown by the ants to suppress the invading fungus, Escovopsis (29).
Results
Controlled-Feeding Studies (TDFglu-phe and TPglu-phe Estimates).
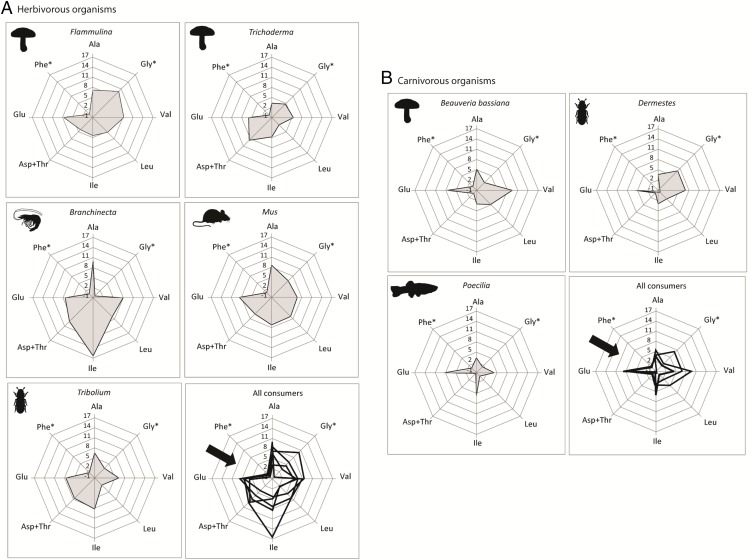
Trophic discrimination factors (15N-discrimination between a consumer and its diet) were measured for specific amino acids extracted from fungal, bacterial, and animal consumers. The degree of 15N-discrimination in glutamic acid (glu) was consistently high among all consumers (7.06 ± 1.64‰), whereas that of phenylalanine (phe) (−0.16 ± 1.55‰) was trivial. This pattern stood in stark contrast to the more variable patterns observed in other amino acids (Table S1), whether feeding on a plant-derived (Fig. 2A) or animal-derived diet (Fig. 2B). Here, the “glu-phe elbow” highlights the value of glu and phe as trophic and source amino acids, respectively. Trophic amino acids (TrAA) enrich significantly between trophic levels, whereas source amino acids (SrcAAs) tend to be insensitive to intertrophic enrichment (17, 21). As such, these two classes of amino acids have served to indicate the TPs of animal taxa (17, 20, 23, 32). We were able to consistently measure the δ15N of the following TrAAs: alanine, glutamic acid, leucine, valine, and isoleucine. Among the SrcAAs, we were able to consistently measure glycine, and phenylalanine (Table S1). Across the three biological kingdoms examined in our study, glu and phe produced the most consistent, least variable TDF value, the parameter critical for accurate trophic position estimation.
Table S1.
Intertrophic 15N-discrimination (δ15N ‰) between consumers and their respective diets
| Trophic amino acids (mean δ15N ‰) | Source amino acids (mean δ15N ‰) | ||||||
| Consumer | ala | glu | leu | val | ile | gly | phe |
| Fungi | |||||||
| Aspergillus* | 6.73 | 8.46 | 3.77 | 4.67 | 3.53 | 8.47 | −0.13 |
| Beauveria† | 4.97 | 7.26 | 4.87 | 8.77 | 2.55 | 2.08 | 0.05 |
| Flammulina‡ | 7.17 | 7.69 | 5.27 | 8.29 | 4.42 | 9.98 | 0.37 |
| Gloeophyllum§ | 11.47 | 8.44 | 6.14 | 10.97 | 7.76 | −3.96 | 0.45 |
| Gloeophyllum¶ | 2.12 | 6.02 | −0.48 | 4.13 | 3.59 | −3.13 | −1.07 |
| Irpex§ | 7.39 | 6.95 | 5.63 | 7.76 | 8.63 | 8.04 | −0.79 |
| Irpex¶ | −3.88 | 2.35 | 0.53 | 1.80 | 4.07 | 4.21 | −5.01 |
| Leucoagaricus‡ | 4.67 | 6.58 | 2.83 | 4.73 | 4.22 | −4.51 | 0.23 |
| Trichoderma‡ | 3.33 | 5.90 | 2.05 | 5.43 | 4.75 | 4.69 | 0.06 |
| Bacteria | |||||||
| Escherichia|| | 6.29 | 6.83 | 1.63 | 3.26 | 0.04 | 1.02 | 0.35 |
| Streptomyces** | 3.35 | 6.97 | 3.13 | 1.97 | 4.81 | 6.99 | 0.15 |
| Insects | |||||||
| Dermestes† | 3.41 | 5.32 | 2.61 | 6.35 | 2.49 | 6.95 | −2.36 |
| Plodia†† | 9.27 | 9.76 | 4.42 | 10.96 | −0.62 | 6.13 | 1.71 |
| Tribolium‡ | 6.51 | 7.57 | 2.01 | 6.22 | 8.28 | 3.11 | 1.26 |
| Crustaceans | |||||||
| Branchinecta‡ | 10.07 | 7.32 | 7.02 | 7.93 | 16.36 | −0.24 | 0.62 |
| Fish | |||||||
| Poecillia† | 3.12 | 7.98 | 0.45 | 3.55 | 5.26 | 1.36 | 0.42 |
| Mammals | |||||||
| Mus‡ | 8.65 | 8.60 | 7.05 | 6.68 | 7.21 | 6.03 | 1.00 |
| Mean | 5.57 | 7.06 | 3.47 | 6.09 | 5.14 | 3.37 | −0.16 |
| SD | 3.62 | 1.64 | 2.31 | 2.79 | 3.87 | 4.45 | 1.55 |
15N-discrimination was measured among the most consistently measured amino acids: alanine (ala), glutamic acid (glu), leucine (leu), valine (val), isoleucine (ile), glycine (gly), and phenylalanine (phe). δ15N values (‰) represent mean enrichment between consumers and their respective diets, derived from at least three samples of each consumer type and diet type.
Consumer taxa were cultured on *cranberry leaves, †fall armyworm, ‡soy-wheat blend, §birch wood, ¶maple wood, ||broth of yeast-extract plus tryptone, **yeast-extract, sucrose, and tryptic soy broth, or ††shiitake mushrooms. Trophic amino acids are a class of compounds thought to enrich substantially with each trophic transfer, whereas source amino acids are thought to change little, mirroring background 15N signatures.
Fig. 2.
Patterns of intertrophic 15N-discrimination within specific amino acids, across a broad diversity of heterotrophic organisms. For each amino acid (AA) extracted from a given consumer, the net change in δ15NAA between the consumer and its diet has been arrayed within a radar plot. This net change is also commonly referred to as the TDF, an important parameter in trophic position estimation. All consumers were fed exclusively on either (A) a plant-based diet (soy and wheat flour) or (B) an animal-based diet. The consistently high degree of 15N-discrimination in glutamic acid (glu) stands in stark contrast to the consistently low discrimination in phenylalanine (phe). When these intertrophic discrimination patterns are superimposed over one another, a distinct “elbow” is formed by the high glu, low phe pattern (creating the “glu-phe elbow,” indicated by bold arrows). This phenomenon is typical of isotopic fractionation in animal tissues (17, 20, 21, 23) and underscores the finding that δ15Nglu and δ15Nphe discrimination patterns in fungi and bacteria mirror those of animals. *Source AAs.
Among the plant-feeding fungi in our controlled-feeding trials, these consumers produced a TDFglu-phe (mean ± 1σ) of 7.26 ± 1.22‰ (Table 1). This value was not significantly different (t30 = 1.01, P = 0.320) from that of our plant-feeding animals (6.79 ± 0.76‰), nor was it significantly dissimilar from the standard TDFglu-phe, +7.64‰ (t23 = −1.38, P = 0.181). Carnivorous fungi produced a mean TDFglu-phe of 7.21 ± 1.29‰, which was not significantly different (t8 = −0.305, P = 0.768) from that of animals fed the same diet (7.43 ± 0.92‰), nor from the standard TDFglu-phe (t2 = −0.575, P = 0.624). Similarly, the bacteria cultured on yeast-extract media produced a mean TDFglu-phe value of 6.65 ± 0.33‰. This degree of enrichment was not significantly different from those of either fungi or animals (F2,45 = 0.803, P = 0.454). Pooling together the bacterial and fungal consumers into a single microbial group, the microbial TDFglu-phe was not dissimilar from that of the animals in our study (t46 = 0.251, P = 0.803).
Table 1.
Comparison of the TDFs and TPs of fungal, bacterial, and animal taxa
| Consumer | Diet type | TDFglu-phe | TDFTrAA-SrcAA | TPexpected | TPglu-phe |
| Fungi | |||||
| Aspergillus | Plant | 8.59 | 0.95 | 2 | 2.1 |
| Beauveria | Animal (herbivore) | 7.21 | 4.62 | 3 | 3.0 |
| Flammulina | Plant | 7.32 | 0.88 | 2 | 2.0 |
| Gloeophyllum (on birch) | Plant | 7.99 | 10.77 | 2 | 2.0 |
| Gloeophyllum (on maple) | Plant | 7.10 | 5.18 | 2 | 1.9 |
| Irpex (on birch) | Plant | 7.74 | 3.65 | 2 | 2.0 |
| Irpex (on maple) | Plant | 7.36 | 1.37 | 2 | 2.0 |
| Leucoagaricus | Plant | 6.35 | 7.12 | 2 | 1.8 |
| Trichoderma | Plant | 5.83 | 1.92 | 2 | 1.8 |
| Bacteria | |||||
| Escherichia | Yeast-extract, tryptone | 6.48 | 3.62 | 3.5 | 3.4 |
| Streptomyces | Yeast extract, tryptic soy | 6.82 | 3.72 | 3.0 | 2.9 |
| Insects | |||||
| Dermestes | Animal (herbivore) | 7.68 | 1.74 | 3 | 3.1 |
| Plodia | Fungus (herbivore) | 8.05 | 4.62 | 3 | 3.0 |
| Tribolium | Plant | 6.31 | 3.93 | 2 | 1.8 |
| Crustaceans | |||||
| Branchinecta | Plant | 6.70 | 9.55 | 2 | 1.9 |
| Fish | |||||
| Poecillia | Animal (herbivore) | 7.56 | 3.18 | 3 | 3.0 |
| Mammals | |||||
| Mus | Plant | 7.59 | 4.12 | 2 | 2.0 |
| Mean TDF | 7.22 | 4.17 | |||
| SD (σ) | 0.73 | 2.79 | |||
| % RSD | 10.06% | 66.83% | |||
Values calculated using glutamic acid and phenylalanine, as well as multicompound groupings of TrAA and SrcAA. TPexpected indicates the expected trophic position of a consumer, given the known trophic position of its diet. TPglu-phe indicates the observed trophic position of each consumer, based on its 15Nglu and 15Nphe values. TrAA = alanine, valine, leucine, isoleucine, glutamic acid. SrcAA = glycine, phenylalanine. The % relative SD (% RSD) quantifies the ratio of the SD to its corresponding mean (×100) and conveys the degree of variability as a proportion of the mean. Among the 17 consumer-diet combinations examined in this study, the TDFTrAA-SrcAA was much more variable than the TDFglu-phe (equal variance test, P < 0.01).
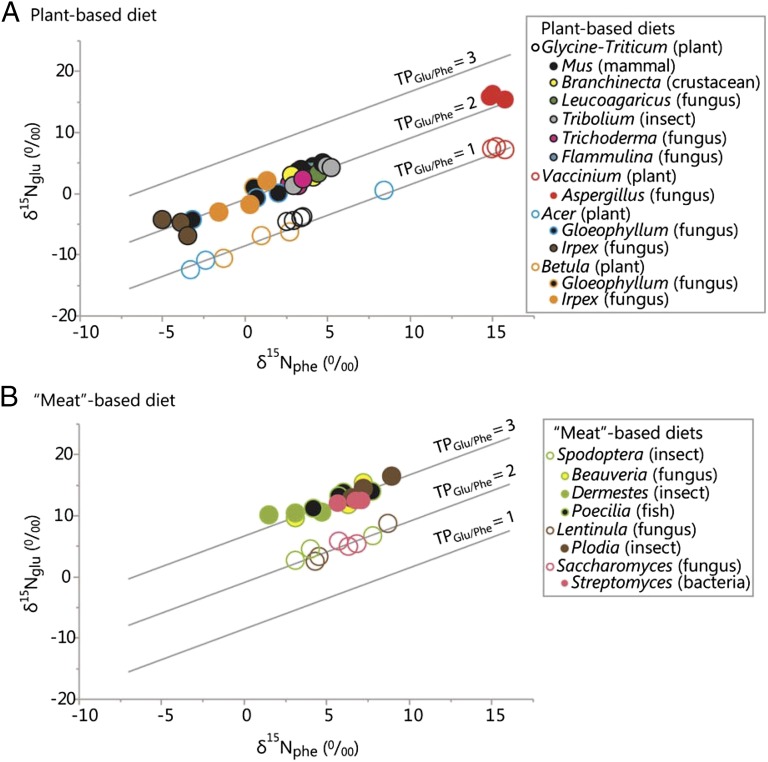
Because our fungal, bacterial, and animal species were kept in pure culture and fed an isotopically characterized, homogeneous diet, their trophic positions were known. We were thus able to compare the known TP of a consumer with its empirically measured TPglu-phe (Table 1). The mean TPglu-phe values of plant-feeding fungi (1.96 ± 0.153) and animals (1.90 ± 0.093) were not different from each other (t31 = 0.97, P = 0.338; Fig. 3A). Similarly, the mean TPglu-phe of fungi (3.01 ± 0.170) and animals (3.04 ± 0.113) feeding exclusively on the animal-based diet were not different from each other (t8 = −0.39, P = 0.710; Fig. 3B). Bacterial trophic positions were examined using diets more suited to bacterial culture (yeast-based growth media). Streptomyces and Escherichia coli were cultured on broths in which yeast extracts (fungus-derived amino acids) comprised the primary protein sources. The yeasts used in these diets (Saccharomyces spp., typically S. cerevisiae) are cultured on plant biomass, thus the yeasts are herbivorous organisms and registered as such: the TPglu-phe of the yeast-based diet registered 2.00 ± 0.075. Streptomyces, the bacterium consuming this diet, registered at 2.91 ± 0.039, which was one trophic level higher and distinctly carnivorous (Table 1). The E. coli diet was a different broth blend consisting of both yeast extract and tryptone (amino acids of unknown origin), and this diet registered a mean TPglu-phe of 2.54 ± 0.025. The TP of E. coli cultured on this diet was 3.40 ± 0.043. Again, the bacterium registered one trophic level higher than its diet.
Fig. 3.
Amino acid isotope compositions (δ15Nglu and δ15Nphe) of fungal and animal species, arrayed across trophic isoclines. (A) Organisms cultured on a plant-based diet. (B) Organisms cultured on an animal-based diet. For each organism, the glutamic acid δ15N value (δ15Nglu) is plotted against its corresponding phenylalanine δ15N value (δ15Nphe). ○, samples of a given diet; ●, consumers. Trophic isoclines (plotted as solid lines) represent pairings of δ15Nglu and δ15Nphe values over a wide gradient, with each line corresponding to an integer trophic position (TP = 1, 2, and 3).
To further test the hypothesis that microbes are trophically equivalent to animals, homogenized diets of fungus-derived proteins were fed to animal and bacterial species. Dried shiitake mushrooms, Lentinula edodes, were fed to the larvae of a moth, Plodia interpunctella. Shiitake mushrooms are wood-eating fungi and registered as herbivores (TPglu-phe = 1.98 ± 0.120). The Plodia larvae feeding on the mushrooms registered as carnivores, with a mean TPglu-phe of 3.04 ± 0.073 (Table 1). As mentioned previously, the bacterium, Streptomyces, was grown on media containing extracts from herbivorous yeast cultures. Streptomyces registered as a carnivore, with a TPglu-phe of 2.91 ± 0.039, approximately one trophic level higher than its yeast-based diet.
The plant-based diets in our study represented TP = 1 and were measured as such (Fig. 3A). When these measurements were arrayed within a δ15N biplot, they closely aligned with the trophic isocline corresponding to primary producers. Trophic isoclines (trophoclines) are pairings of δ15Nglu and δ15Nphe values that, when plotted across a wide gradient of δ15N signatures, represent the integer TPs within a food chain (e.g., TP = 1, 2, or 3). The herbivores in our controlled-feeding trials (known TP = 2) were closely aligned with trophocline 2 (Fig. 3A), and the carnivores (known TP = 3) were all clustered along trophocline 3 (Fig. 3B). Among all fungal, bacterial, and animal taxa, the observed TPglu-phe values were not significantly different from their respective, known TPs (n = 17, paired Wilcoxon signed rank test: W = −31.00, P = 0.074).
Fungus-Garden Study.
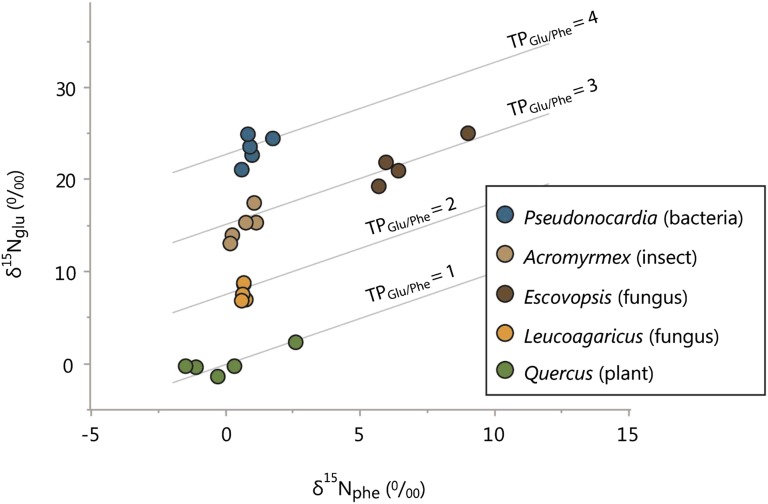
An examination of leaf-cutter ant fungus gardens (Fig. 1) revealed that the ant-cultivated fungus, Leucoagaricus, was an herbivorous organism (TPglu-phe = 1.9 ± 0.12), whereas the ants, long known to be fungivorous (28), registered at a distinctly carnivorous TP: 2.9 ± 0.17 (Fig. 4). The invading fungus, Escovopsis, also registered at a carnivorous TP, 3.0 ± 0.15, and the bacterium deployed by the ants to defend the fungus garden registered a TP of 4.0 ± 0.17. Arrayed across trophoclines, the bacterial, fungal, and animal consumer groups within the fungus garden registered as four discrete trophic groups (Fig. 4).
Fig. 4.
Amino acid isotope compositions (δ15Nglu and δ15Nphe) of each trophic group within ant fungus gardens. For each organism, the glutamic acid δ15N value (δ15Nglu) is plotted against its corresponding phenylalanine δ15N value (δ15Nphe). All isotopic values (δ15N) are arrayed across trophic isoclines. Trophic isoclines (plotted as solid lines) represent pairings of δ15Nglu and δ15Nphe values over a wide gradient, with each line corresponding to an integer trophic level (TPglu/phe = 1–4).
Discussion
Viewed under the lens of amino acid isotopic analysis, the bacteria, fungi, and animals in our study exhibited strikingly similar patterns of intertrophic 15N-discrimination. Specifically, the consistent TDFglu-phe among all consumers reflected predictable patterns of 15N-discrimination in glutamic acid and phenylalanine, the two amino acids previously shown to be critically important to accurate TP estimation among animals (17, 20, 26, 33). The degree of intertrophic 15N-discrimination in glu was relatively high, whereas that of phe was characteristically low, which contrasted with the more variable patterns observed in the remaining amino acids. This glu-phe 15N-discrimination pattern held true across not only a broad phylogenetic spectrum (three biological kingdoms), but also a diversity of ecosystem types (terrestrial vs. aquatic) and trophic groups (plant- vs. animal-based diets). Trophically, therefore, the macro- and microfauna in our study were equivalent. Such constancy in the TDFglu-phe facilitates accurate, predictable trophic position estimation within the broader empire of heterotrophy.
The degree to which microbial and animal species can be integrated within a food web is particularly apparent when δ15Nglu and δ15Nphe values are arrayed across trophoclines. Trophoclines effectively couple δ15Nglu and δ15Nphe measurements within a 2D isotopic space and thereby provide a framework in which to view consumer trophic position. Within this framework, our microbial taxa registered as strict carnivores (trophic position 3) when feeding on herbivores, as did our animal taxa. Importantly, the animal taxa feeding on the same or analogous resources were trophically indistinguishable from the microbes, permitting the interdigitation of macro- and microfauna along each trophocline. Here, it is apparent that the consumers in our study registered one trophic level higher than their respective diets, regardless of consumer identity. Thus, whether it was crustaceans, fungi, insects, or mammals feeding on plant-based diets, the consumers registered as herbivores. Likewise, whether we measured moths eating fungi, fish eating homogenized insect powder, bacteria consuming yeast extracts, or fungi eating caterpillars, the consumer taxa registered as strict carnivores within the trophic hierarchy.
These findings suggest that when organisms feed on microbes, the consumers’ trophic positions elevate predictably, regardless of whether the consumer is an animal or microbe. Trophically, therefore, a heterotrophic microbe represents “meat” in a food web. In light of the predominance of microbial detritivores within brown food webs (7, 12, 13), microbial biomass may be a profoundly important source of protein flowing up through food chains. Indeed, as food chains shorten with the trophic downgrading of ecosystems (5), higher-order microbial carnivores may provide an important stabilizing buffer against the asymmetries caused by the loss of other trophic groups. Future work in this vein might investigate whether microbes and animals commonly swap trophic roles when the other is lost.
Within leaf-cutter ant fungus gardens, we examined the trophic roles of each symbiont in the community. Using amino acid stable isotope fingerprinting, we show that the cultivated fungus, Leucoagaricus, fed as an herbivore (TPglu-phe = 1.9 ± 0.12) and was the sole consumer of plant material in the fungus gardens. That a fungus was the sole herbivore within the fungus-garden community suggests that fungi, not ants, are the dominant herbivores of the Neotropics. The ants, long known to be fungivorous (28), registered at a distinctly carnivorous TPglu-phe: 2.9 ± 0.17. Importantly, the TPglu-phe of the ants was exactly one trophic level above their diet, and as strict consumers of herbivores, the ants were functional carnivores. In the trophic hierarchy of a fungus garden, the ants hold an intermediate position (trophic level 3) and thus are more analogous to “ranchers” than “gardeners.” The invading fungus, Escovopsis, also registered at a carnivorous TPglu-phe, 3.0 ± 0.15, indicating that it, too, fed on the herbivores of the community. At this TPglu-phe, Escovopsis appears to be a direct competitor of the ants and not a consumer of the ants. To better compete with Escovopsis, the ants deploy their bacterial symbiont, Pseudonocardia, and interestingly, this bacterium registered a TPglu-phe of 4.0 ± 0.17, one trophic level above the ants. Given that the bacterium grows on the ant exoskeleton (Fig. 1D) and is closely associated with specialized glands within cuticular crypts (29), the observed trophic position of Pseudonocardia strongly suggests it feeds exclusively on the ants. The tradeoff underlying the ant-bacterial mutualism, therefore, can be characterized as food for protection, wherein the ants use their own tissues to culture a carnivorous bacterium, and in exchange, the bacterium protects the fungus garden from invaders. Collectively, these findings reveal the presence of four discrete trophic levels within the fungus garden community. Here, the bacterium is the trophic equivalent of an apex carnivore, whereas the ant colony and its fungal competitor feed as meso-carnivores, and all are supported by the foundational herbivore of the community, another fungus. By coupling isotopic fingerprinting with natural history information, we are able to better illuminate the trophic identities of fungal, bacterial, and animal consumers in an ancient, quadripartite symbiosis.
Our overarching goal in this work is to provide a basis to accurately interpret the trophic positions of free-roaming heterotrophic organisms, regardless of their phylogenetic origin or ecosystem type. We provide empirical evidence that isotopically derived trophic metrics are equally applicable to animal and microbial consumer groups. Whether feeding on plant- or animal-based biomass, fungi and bacteria are trophic analogs of animals. Although it is possible that the 15 taxa cultured in our controlled-feeding studies were not broadly representative of heterotrophic bacteria, fungi, and animals, the likelihood that we inadvertently selected the 15 anomalous species is quite small given the exceedingly high global diversity of heterotrophic fauna (3, 4, 7, 8). It is more parsimonious to conclude that the organisms in our study reflect real patterns within the broader heterotrophic empire and that microbes can be considered the trophic equivalents of animals within a food chain. For food web ecology, this reframes how the microbiome can be viewed and resolves long-standing questions as to where microbes fit within the food chain. Fungal, bacterial, and animal species can be integrated within a single trophic hierarchy, thereby uniting the macro- and microbiome and facilitating more comprehensive assessments of functional diversity within ecosystems.
Materials and Methods
Culturing of Consumer Species.
Fifteen heterotrophic species, spanning six phyla (Proteobacteria, Actinobacteria, Ascomycota, Basidiomycota, Arthropoda, and Chordata) and three kingdoms (Fungi, Bacteria, and Animalia), were kept in pure culture on homogeneous, isotopically characterized diets (SI Materials and Methods). Among the animal taxa cultured were crustaceans, fish, insects, and mammals. Among the microbiota, we cultured ascomycete and basidiomycete fungi, as well as proteobacteria and filamentous bacteria (actinomycetes). When a consumer had developed to maturity on a given diet, the entire organism was homogenized and the δ15N values of amino acids within its tissues were measured using established analytical protocols (17, 26, 34). Cultures of fungus gardens were maintained within a controlled-environment laboratory setting (24–26 °C, 16:8 photoperiod). Leaf-cutter ant (Acromyrmex echinatior) colonies were provisioned with leaves harvested from oak trees (Quercus macrocarpa). Colonies were confined to clear plastic mesocosms and maintained according to established rearing protocols (29).
Compound-Specific Isotope Analysis.
Each specimen was collected, euthanized, and desiccated in a drying oven for 7–14 d and then homogenized before drawing aliquots for stable nitrogen isotope analysis of amino acids. Compound-specific isotope analysis of N was conducted via protocols (17, 34) developed and refined at the Department of Biogeochemistry, Japan Agency of Marine-Earth Science and Technology (JAMSTEC), Yokosuka, Japan. In brief, specimens were hydrolyzed and derivatized, allowing for the extraction of amino acids. The identities of the amino acids were verified via gas chromatography (GC) and then combusted (C) within a furnace interfaced with an isotopic ratio MS (IRMS). Using an integrated GC-C-IRMS system, each target amino acid had its isotope ratio quantified independently. The δ15N values were determined for a suite of amino acids, generally including alanine, glutamic acid, leucine, valine, isoleucine, glycine, and phenylalanine (Datasets S1–S30).
Trophic Computations and Statistics.
Trophic position (TPglu-phe) estimates were generated using the following equation (17):
| [1] |
where δ15Nglu represents the nitrogen isotopic ratio of glutamic acid, δ15Nphe represents the nitrogen isotopic ratio of phenylalanine, β corrects for the difference in 15N values between glutamic acid and phenylalanine within the primary producers of the food web (e.g., β ∼ 8.4‰ for C3 plants), Δglu-phe represents the net trophic discrimination between glutamic acid and phenylalanine, and λ represents the basal trophic level (=1) of the food web. The trophic discrimination factor, Δglu-phe (also referred to as the TDF), represents the net intertrophic 15N-discrimination between glutamic acid and phenylalanine (SI Materials and Methods). Discernment of significant differences between known and observed TP values was examined using univariate ANOVA and nonparametric tests (paired Wilcoxon signed rank tests where data were heteroscedastic). Distinguishing among TDF values was accomplished using paired t tests.
SI Materials and Methods
Trophic Computations.
The δ15N value for any given sample is determined via the following: δ15N = [(Rsample/Rstandard) – 1)] × 1,000, where Rsample represents the 15N:14N ratio in a sample, and Rstandard represents the 15N:14N ratio of atmospheric nitrogen (N2). The unit of measurement for δ15N is per-mil (‰).
In Eq. 1, the trophic discrimination factor (Δglu-phe) represents the net 15N-discrimination between trophic levels and is often measured as the net intertrophic discrimination among two particular amino acids: glutamic acid and phenylalanine (17, 20). The net trophic discrimination factor is calculated as the following:
| [S1] |
This enrichment between a consumer and its diet occurs as dietary proteins are deaminated and/or transaminated within the consumer, allowing for physiological discrimination among molecules bearing heavy or light isotopes (17, 22). Across a broad range of trophic levels, taxa, and ecosystem types, Δglu-phe (TDFglu-phe) has been shown to be approximately +7.64 ‰ (17, 19, 20, 22).
Eq. 1 can be reconfigured algebraically to provide the equation for a trophic isocline (trophocline) at any given integer trophic position (e.g., TP = 1, 2, 3)
| [S2] |
These two amino acids (glu and phe) vary independently, but importantly, the relationship between them remains constant across taxa, trophic levels, and ecosystem types (20, 22). Thus, given any δ15Nphe value, there is a δ15Nglu value corresponding to a designated integer trophic level (e.g., trophic levels 1, 2, or 3). Over a wide δ15N gradient, these δ15Nphe and δ15Nglu pairings produce trophoclines that clarify trophic position against widely varying background δ15N signatures (26).
Culturing of Consumer Species.
To create discrete trophic groups of known trophic position, fungal, bacterial, and animal consumers were reared to maturity in either Petri dishes or glass mesocosms under standard laboratory protocols (20, 28, 29). Rearing durations ranged from 3 wk to 7 mo, depending on the biology of the organism. Among the animal taxa cultured were crustaceans (fairy shrimp, Branchinecta), fish (common guppies, Poecilia reticulata), insects (red flour beetles, Tribolium castaneum; pantry moths, Plodia interpunctella; skin beetles, Dermestes), and mammals (common house mice, Mus musculus). Among the fungi, we cultured species from the Ascomycota (black mold, Aspergillus niger; green mold, Trichoderma viride; white muscardine, Beauveria bassiana) and the Basidiomycota (enoki mushrooms, Flammulina velutipes; brown-rot wood fungus, Gloeophyllum trabeum; white-rot wood fungus, Irpex lacteus; leaf-cutter ant fungal symbiont, Leucoagaricus gongylophorus). Among the bacteria, two species were cultured: an actinomycete in the genus Streptomyces (sirexAA-E) and a proteobacterium, Escherichia coli (DH5-α). Importantly, each fungal, bacterial, and animal consumer was a unique replicate of heterotrophy; thus, our experimental design allowed for our inference to be generalized across the broader pool of heterotrophy.
Consumer taxa were fed either plant- or animal-based biomass (Table S1). There were four plant-based diets, including two herbaceous plants, two trees, and a woody vine. The herbaceous diet was a blend of soy bean (Glycine max) powder (25% by volume) and wheat (Triticum aestivum) flour (75% by volume). The biomass of two trees, maple (Acer) and birch (Betula), served as the tree-based diets. Leaves of the ericaceous vine, cranberry (Vaccinium macrocarpon) served as the fourth diet. To facilitate the separation of diet from consumer, the bacterial species in our study were cultured on yeast-extract broths. E. coli was grown on a broth composed of water, yeast extract, tryptone, and NaCl. Streptomyces was grown on tryptic soy broth, yeast extract, and sucrose. The animal-based diet was a dried, powdered homogenate of caterpillars (larval Spodoptera frugiperda). Fungi alone (shiitake mushrooms, dried) were used to feed Plodia larvae.
Selection of consumer taxa was based on currently available cultures, the capacity of a given species to feed entirely on a dried food source, and the availability of vertebrate colonies at University of Wisconsin rearing facilities. It was particularly important to establish cultures of animals that could feed exclusively on dried biomass (e.g., Plodia moths, Dermestes beetles, and Tribolium beetles) because of the ubiquity and omnipresence of microbial detritivores in the diet itself. A food source that has partially decomposed before an animal feeds on it will have a distinctly different isotopic value because the microbes have literally consumed much of the dietary protein already (and fractionated the isotopes in both their own biomass and that of the diet).
Supplementary Material
Acknowledgments
We thank Janet van Zoeren, Rachel Arango, Sacha Horn, Lindsay Wells, Brian Hudelson, Christopher Watson, Merritt Singleton, and Drs. Patricia McManus, Bhadriraju Subramanyam, Tess Killpack, and Bill Karasov for assistance with animal and microbial cultures. Leaf-cutter ant photos appear courtesy of Don Parsons. Drs. Prarthana Dharampal, Peggy Ostrom, Stephen Carpenter, and Elissa Chasen provided helpful suggestions on earlier manuscript drafts. This work was supported by the University of Wisconsin Vilas Lifecycle Professorship (awarded to S.A.S.), the US Department of Agriculture–Agricultural Research Service (Current Research Information System 3655-21220-001, awarded to S.A.S. and J.E.Z.), and the Japan Agency for Marine-Earth Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508782112/-/DCSupplemental.
References
- 1.Chase J, Leibold M. Ecological Niches: Linking Classical and Contemporary Approaches. Univ of Chicago Press; Chicago: 2003. [Google Scholar]
- 2.Cohen J, et al. Improving food webs. Ecology. 1993;74(1):252–258. [Google Scholar]
- 3.May RM. How many species are there on Earth? Science. 1988;241(4872):1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- 4.Pimm SL, et al. The biodiversity of species and their rates of extinction, distribution, and protection. Science. 2014;344(6187):1246752. doi: 10.1126/science.1246752. [DOI] [PubMed] [Google Scholar]
- 5.Estes JA, et al. Trophic downgrading of planet Earth. Science. 2011;333(6040):301–306. doi: 10.1126/science.1205106. [DOI] [PubMed] [Google Scholar]
- 6.Hooper D, et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol Monogr. 2005;75(1):3–35. [Google Scholar]
- 7.Beattie A, Ehrlich P. The missing link in biodiversity conservation. Science. 2010;328(5976):307–308. doi: 10.1126/science.328.5976.307-c. [DOI] [PubMed] [Google Scholar]
- 8.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67(10):4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polis G, Strong D. Food web complexity and community dynamics. Am Nat. 1996;147(5):813–846. [Google Scholar]
- 10.Lindeman R. The trophic-dynamic aspect of ecology. Ecology. 1942;23(4):399–417. [Google Scholar]
- 11.Kaspari M, Yanoviak SP. Biogeochemistry and the structure of tropical brown food webs. Ecology. 2009;90(12):3342–3351. doi: 10.1890/08-1795.1. [DOI] [PubMed] [Google Scholar]
- 12.Coleman DC. Energetics of detritivory and microbivory in soil in theory and practice. In: Polis GA, Winemiller KO, editors. Food Webs: Integration of Patterns and Dynamics. Chapman & Hall; New York: 1996. pp. 39–50. [Google Scholar]
- 13.Moore JC, de Ruiter PC. Energetic Food Webs. Oxford Univ Press; Oxford, UK: 2012. [Google Scholar]
- 14.Colinvaux P. Why Big Fierce Animals Are Rare: An Ecologist’s Perspective. Princeton Univ Press; Princeton: 1978. [Google Scholar]
- 15.Haraguchi TF, Uchida M, Shibata Y, Tayasu I. Contributions of detrital subsidies to aboveground spiders during secondary succession, revealed by radiocarbon and stable isotope signatures. Oecologia. 2013;171(4):935–944. doi: 10.1007/s00442-012-2446-1. [DOI] [PubMed] [Google Scholar]
- 16.Tayasu I, Hyodo F. Use of carbon—14 natural abundances in soil ecology: Implications for food web research. In: Ohkouchi N, Tayasu I, Koba K, editors. Earth, Life, and Isotopes. Kyoto Univ Press; Kyoto: 2010. pp. 3–16. [Google Scholar]
- 17.Chikaraishi Y, et al. 2009. Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol Oceanogr 7(2003):740–750.
- 18.Hannides C, Popp B, Landry M, Graham B. Quantification of zooplankton trophic position in the North Pacific Subtropical Gyre using stable nitrogen isotopes. Limnol Oceanogr. 2009;54(1):50–61. [Google Scholar]
- 19.Chikaraishi Y, Ogawa NO, Doi H, Ohkouchi N. 15N/14N ratios of amino acids as a tool for studying terrestrial food webs: A case study of terrestrial insects (bees, wasps, and hornets) Ecol Res. 2011;26(4):835–844. [Google Scholar]
- 20.Steffan SA, et al. Trophic hierarchies illuminated via amino acid isotopic analysis. PLoS One. 2013;8(9):e76152. doi: 10.1371/journal.pone.0076152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClelland J, Montoya J. Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology. 2002;83(8):2173–2180. [Google Scholar]
- 22.Chikaraishi Y, Steffan SA, Takano Y, Ohkouchi N. Diet quality influences isotopic discrimination among amino acids in an aquatic vertebrate. Ecol Evol. 2015;5(10):2048–2059. doi: 10.1002/ece3.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popp B, et al. Insight into the trophic ecology of yellowfin tuna, Thunnus albacares, from compound‐specific nitrogen isotope analysis of proteinaceous amino acids. In: Dawson T, Siewolf R, editors. Stable Isotopes as Indicators of Ecological Change. Academic; London: 2007. pp. 173–190. [Google Scholar]
- 24.Newsome SD, Fogel ML, Kelly L, Martínez del Rio C. Contributions of direct incorporation from diet and microbial amino acids to protein synthesis in Nile tilapia. Funct Ecol. 2011;25(5):1051–1062. [Google Scholar]
- 25.Maraun M, et al. Stable isotopes revisited: Their use and limits for oribatid mite trophic ecology. Soil Biol Biochem. 2011;43(5):877–882. [Google Scholar]
- 26.Chikaraishi Y, et al. High-resolution food webs based on nitrogen isotopic composition of amino acids. Ecol Evol. 2014;4(12):2423–2449. doi: 10.1002/ece3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerling T, Harris J. Carbon isotope fractionation between diet and bioapatite in ungulate mammals and implications for ecological and paleoecological studies. Oecologia. 1999;120(3):347–363. doi: 10.1007/s004420050868. [DOI] [PubMed] [Google Scholar]
- 28.Currie C, Scott J, Summerbell R, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–705. [Google Scholar]
- 29.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311(5757):81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 30.Hölldobler B, Wilson EO. The Ants. Belknap Publishing; Cambridge, MA: 1990. [Google Scholar]
- 31.Currie CR, et al. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003;299(5605):386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 32.Décima M, Landry MR, Popp BN. Environmental perturbation effects on baseline δ15N values and zooplankton trophic flexibility in the southern California Current Ecosystem. Limnol Oceanogr. 2013;58(2):624–634. [Google Scholar]
- 33.Chikaraishi Y, Kashiyama Y, Ogawa NO, Kitazato H, Ohkouchi N. 2007. Metabolic control of nitrogen isotope composition of amino acids in macroalgae and gastropods: Implications for aquatic food web studies. Mar Ecol Prog Ser 342(2003):85–90.
- 34.Chikaraishi Y, Takano Y, Ogawa NO, Ohkouchi N. Instrumental optimization for compound-specific nitrogen isotope analysis of amino acids by gas chromatography/combustion/isotope ratio mass spectrometry. In: Ohkouchi N, Tayasu I, Koba K, editors. Earth, Life, and Isotopes. Kyoto Univ Press; Kyoto: 2010. pp. 367–386. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.