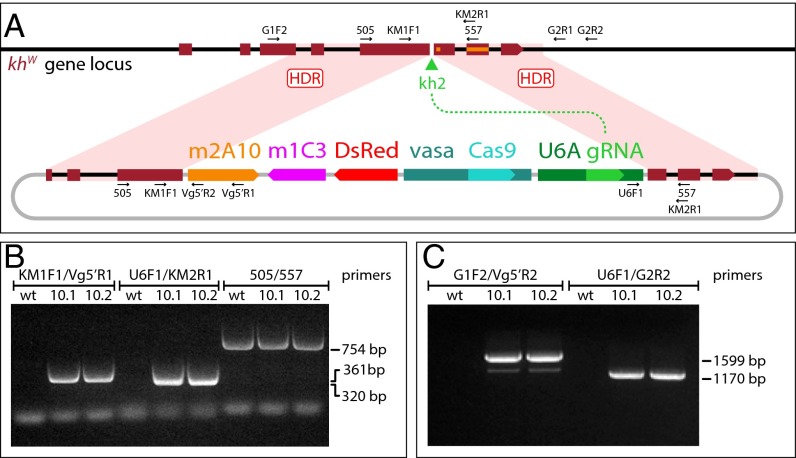
Fig. 1.
Site-specific integration into the An. stephensi kynurenine hydroxylasewhite locus of the gene-drive construct AsMCRkh2, carrying antimalarial effector genes. (A) Schematic representations of the kynurenine hydroxylasewhite locus and AsMCRkh2 construct. Genes and other features of the AsMCRkh2 construct are not to scale. The dark red boxes represent the eight exons of the endogenous khw gene locus (Top) with the direction of transcription indicated by the wedge in exon 8. The black lines represent genomic and intron DNA. The green arrowhead represents the target site of the gRNA, kh2. Labels and arrows indicate names, approximate positions, and directions of oligonucleotide primers used in the study. khw gene sequences corresponding to previously characterized mutations are indicated as an orange rectangle (28) and square (29). The plasmid, AsMCRkh2 (Bottom), carries promoter and coding sequences comprising vasa-Cas9 and the U6A-kh2 gRNA genes (U6A gRNA) linked to the dual scFv antibody cassette (m2A10-m1C3) conferring resistance to P. falciparum (11) and the dominant eye marker gene (DsRed) inserted between regions of homology (dark red boxes) from the An. stephensi khw locus that directly abut the U6A-kh2 gRNA cut site. The black lines represent khw intron sequences, and the gray lines indicate plasmid DNA sequences. Following gRNA-directed cleavage by the Cas9–kh2 gRNA nuclease complex at the kh2 target site (green arrowhead), homology-directed repair (HDR) leads to precise insertion of the AsMCRkh2 cargo (m2A10-m1C3, DsRed, vasa-Cas9, U6A gRNA) into the genomic khw locus via HDR events somewhere within the regions of homology (pink-shaded quadrilaterals). Plasmid sequences are not integrated. (B) Gene amplification analysis confirms integration of the AsMCRkh2 cargo in genomic DNA prepared from the two G1 male transformants (10.1 and 10.2) that were positive for the DsRed eye-marker phenotype. Both males carry left and right junction fragments of the AsMCRkh2 cargo with the supplied khw regions of homology (KM1F1/Vg5′R1 and U6F1/KM2R1 primer combinations, respectively). An amplicon corresponding to the wild-type khw locus (505/557 primer pairs) confirms that these mosquitoes were heterozygous in some of their cells. Wild-type (wt) control DNA supports amplification only of the wild-type khw locus (505/557 primers). (C) Gene amplification analysis confirms site-specific integration of the AsMCRkh2 construct at the khw locus using primers located outside of the genomic sequence included in the AsMCRkh2 cassette (the left integration junction fragment amplified with primers G1F2/Vg5′R2, and the right junction fragment amplified with primers U6F1/G2R2). Wild-type control DNA did not support amplification of these hybrid fragments. Numbers refer to the length in nucleotides of the amplified fragments. Amplicon primary structure was verified by DNA sequencing (SI Appendix, Fig. S1).