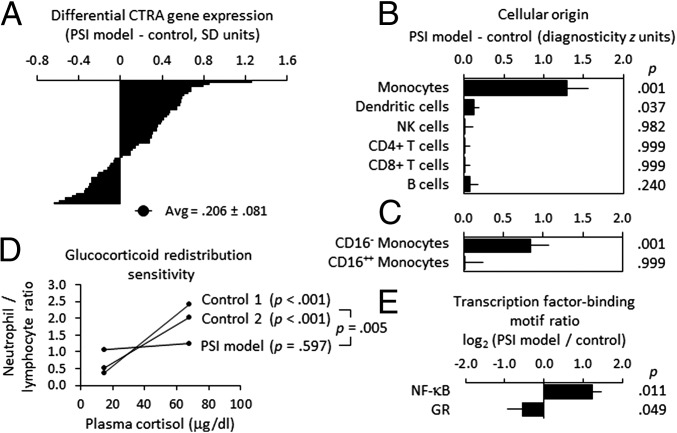
Fig. 3.
Differential gene expression, monocyte transcriptome representation, and glucocorticoid regulation in the rhesus macaque model of PSI. (A) Differential expression of 53 CTRA indicator transcripts in PBMC samples from 49 rhesus macaques classified by behavioral observations as low or intermediate PSI (n = 22 and 19, respectively) vs. high PSI (n = 8) as in Fig. 1A (P = 0.017, controlling for age). (B) Transcript origin analyses (12) identifying major leukocyte subset origins of the 229 gene transcripts that showed ≥1.2-fold differential expression in high-PSI vs. intermediate- and low-PSI animals (70 up-regulated transcripts and 159 down-regulated transcripts are listed in Table S1). (C) Transcript origin analyses assessing the extent to which CD14++/CD16− classical and CD14+/CD16+ nonclassical monocyte transcriptomes contributed to differential gene expression. (D) Functional glucocorticoid sensitivity of myeloid lineage immune cells (neutrophils) was assessed using an in vivo hematological redistribution assay that quantified the extent to which circulating neutrophil numbers (normalized to lymphocyte numbers to control for variations in total white blood cell count) varied as a function of endogenous physiological variations in cortisol levels (22, 24). Log-transformed neutrophil/lymphocyte ratios were subject to mixed effect linear model analysis of four repeated measurements collected at 9:00 AM and 3:00 PM on 2 d from low-PSI animals (n = 172 observations on 27 animals, 16 of whom were also resampled 1 y later), intermediate-PSI animals (n = 132 observations on 21 animals, 12 of whom were resampled 1 y later), and high-PSI animals (n = 32 observations on 6 animals, 2 of whom were resampled 1 y later). Average values are displayed in raw ratio units to facilitate interpretation, and lines span the observed range of cortisol values. (E) In vivo transcriptional activity of glucocorticoid receptor (GR) and NF-κB transcription factors as assessed by TELiS bioinformatic analysis (50) of transcription factor-binding motif prevalence in promoter DNA sequences of genes showing ≥1.2-fold differential gene expression in high-PSI animals vs. low- and intermediate-PSI controls.