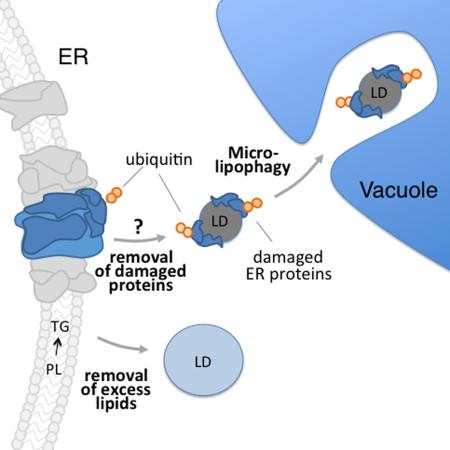
Abstract
The immediate responses to inhibition of phosphatidylcholine (PC) biosynthesis in yeast are altered phospholipid levels, slow growth, and defects in the morphology and localization of ER and mitochondria. With chronic lipid imbalance, yeast adapt. Lipid droplet (LD) biogenesis and conversion of phospholipids to triacylglycerol are required for restoring some phospholipids to near wild-type levels. We confirmed that the Unfolded Protein Response is activated by this lipid stress and find that Hsp104p is recruited to ER aggregates. We also find that LDs form at ER aggregates, contain polyubiquitinated proteins and an ER chaperone, and are degraded in the vacuole by a process resembling microautophagy. This process, microlipophagy, is required for restoration of organelle morphology and cell growth during adaptation to lipid stress. Microlipophagy does not require ATG7 but does requires ESCRT components and a newly identified class E VPS protein that localizes to ER and is up-regulated by lipid imbalance.
Graphical Abstract
Introduction
Defects in the biosynthesis of complex phospholipids are linked to 14 disorders which affect the central and peripheral nervous systems, as well as cardiac and skeletal muscle (Lamari et al., 2013). Recently, a megaconial muscular dystrophy was identified that is due to mutation of the beta isoform of choline kinase, the enzyme that catalyzes the first step in phosphatidylcholine (PC) biosynthesis (Mitsuhashi et al., 2011). Patients with this disease exhibit muscle wasting and weakness from early infancy, ambulatory delays, cognitive disabilities and cardiomyopathy. However, the mechanisms underlying defects in skeletal, cardiac and neurological systems in this disease are not understood. We used the budding yeast Saccharomyces cerevisiae to study the cellular response to defects in the synthesis of PC.
PC biosynthesis occurs via two conserved pathways. One pathway, the phosphatidylethanolamine N-methyltransferase (PEMT) pathway, occurs at sites of close contact between mitochondria and ER (Gaigg et al., 1995; Rusinol et al., 1994). In this pathway, phosphatidylserine (PS) is created from CDP-diacylglycerol and L-serine in the ER (Vance and Steenbergen, 2005) and transported to mitochondria, where it is decarboxylated to form phosphatidylethanolamine (PE) (Voelker, 1997). PE is then transported back to the ER and methylated three times, converting it to PC (Kanipes and Henry, 1997). However, when choline is available, PC is synthesized through the Kennedy pathway. In this pathway, phosphocholine is added to diacylglycerol in a cytidylyltransferase-catalyzed reaction (Gibellini and Smith, 2010).
Previous studies support a link between PC biosynthesis and ER and mitochondria. For example, perturbation of PC biosynthesis results in defects in ER morphology in CHO cells (Testerink et al., 2009) and in mitochondrial morphology and distribution in skeletal muscle fibers of patients with megaconial muscular dystrophy (Mitsuhashi et al., 2011). PC biosynthetic and lipid saturation defects also result in up-regulation of the unfolded protein response (UPR) in yeast, a pathway that is activated by accumulation of unfolded proteins in the ER lumen, and promotes cell survival by inhibiting protein synthesis and promoting refolding or degradation of misfolded proteins (Surma et al., 2013; Thibault et al., 2012). Interestingly, the UPR regulator that localizes to the ER, Ire1p, responds to lipid membrane composition through direct interactions with its transmembrane domain (Volmer et al., 2013). Finally, there are negative genetic interactions between the PC biosynthetic genes (CHO2 and OPI3) and genes involved in mitochondrial inheritance (MMR1, NUM1) or ER protein folding and stress response (EMC complex, GET complex, ERAD proteins and HAC1) in yeast (Costanzo et al., 2010; Hoppins et al., 2011; Surma et al., 2013).
Here, we describe profound defects in yeast growth and organelle organization in response to acute lipid imbalance resulting from defects in PC biosynthesis. We obtained evidence that yeast adapt to lipid imbalance, and a role for lipid droplets (LDs) in that process. LDs consist of a neutral lipid core surrounded by a protein-containing phospholipid monolayer and are produced at ER membranes (Walther and Farese, 2009). We find that LD biogenesis and selective autophagy contribute to the adaptive response by removing excess lipids and damaged proteins from the ER. We also identified a role for a previously uncharacterized open reading frame in control of LD autophagy and adaptation to lipid stress.
Results
Yeast adapt to lipid imbalance produced by defects in PC biosynthesis
Cho2p catalyzes the first step in the conversion of PE to PC in the PEMT pathway for PC biosynthesis (Kanipes and Henry, 1997). Yeast cho2Δ cells can produce PC by the Kennedy pathway when choline is available (Carman and Henry, 1999). We took advantage of this redundancy to examine the effect of short- and long-term PC deficiency in eukaryotic cells. We propagated wild-type and cho2Δ cells in the presence of choline and compared the effects of acute and chronic defects of PC biosynthesis by shifting cho2Δ cells to choline-free media for 1 and 7 days, respectively (Fig. 1, Fig. S1). Thus, cho2Δ+C refers to a cho2Δ cell grown in the presence of choline, while cho2Δ−C1 and cho2Δ−C7 denote cho2Δ strains grown in choline-free media for 1 day (acute) and 7 days (chronic), respectively.
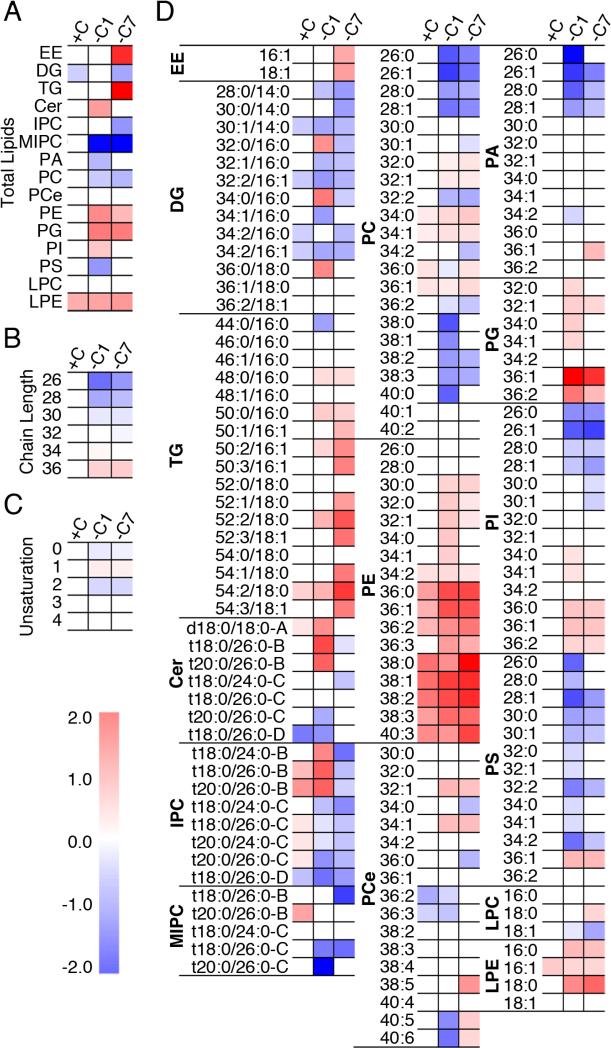
Fig. 1. Phospholipids and neutral lipids change in response to PC biosynthetic defects.
The lipidome of mid-log phase yeast. Heat maps show significant changes using two-way ANOVA followed by post hoc Dunnett's test in (a) lipid subclasses, (b) total fatty acyl/alkyl chain length, (c) unsaturation in phospholipids and DG, and (d) individual lipid species. The color scale indicates log2-transformed fold change of cho2Δ cells vs. WT cells in each condition. See also Figure S1.
The lipid subclass profiles of wild-type (WT) and choline-supplemented cho2Δ (cho2Δ+C) cells are similar. Propagation of cho2Δ cells in the absence of choline for 1 day (cho2Δ−C1) results in decreased levels of PC, phosphatidic acid (PA), PS and myoinositol phosphorylceramide (MIPC), and elevated levels of PE, phosphatidylglycerol (PG), phosphatidylinositol (PI), and ceramide (Cer) compared to wild-type cells (Fig. 1a; Fig. S1b). It also results in an overall increase in the length of fatty acid side chains and monounsaturated side chains compared to wild-type cells (Fig. 1b-c; Fig.S1c-d). Although total levels of triacylglycerol (TG) and ergosteryl ester (EE) do not change during acute lipid imbalance, the lipid species heatmap (Fig. 1d) and/or lipid subclass profiles (Fig. S1b) revealed that some TG and EE species do increase during acute lipid imbalance. This is the first description of the lipidome of yeast experiencing acute defects in PC biosynthesis.
During chronic lipid stress (cho2Δ−C7), the imbalance in PG, PC and MIPC persists, the levels of inositol phosphorylceramide (IPC) decrease, and there is a large increase in the levels of TG and EE (Fig. 1a, 1d). Interestingly, the yeast lipidome adapts: with long-term defects in PC biosynthesis the levels of Cer, PE, PI, PA, and PS are either partially or completely restored to wild-type levels.
PC imbalance triggers defects in mitochondria and ER and in cell growth rates
Using fluorescently tagged mitochondrial and ER marker proteins (citrate synthase (Cit1p) and Pho88p), we studied the effect of PC imbalance on the morphology and distribution of mitochondria and ER, the organelles where PC biosynthesis via the PEMT pathway occur. In wild-type cells, mitochondria are resolved as tubular structures that align along the mother-bud axis and accumulate at the tips of mother cells and buds (Swayne et al., 2011). ER is present as the outer membrane of the nuclear envelope (nuclear ER, nER). It also consists of ER sheets and tubules in the cytosol, as well as cortical ER (cER), a fenestrated network of sheets and tubules that lie beneath and are anchored to the plasma membrane (PM) (Fig. 2a) (Manford et al., 2012).
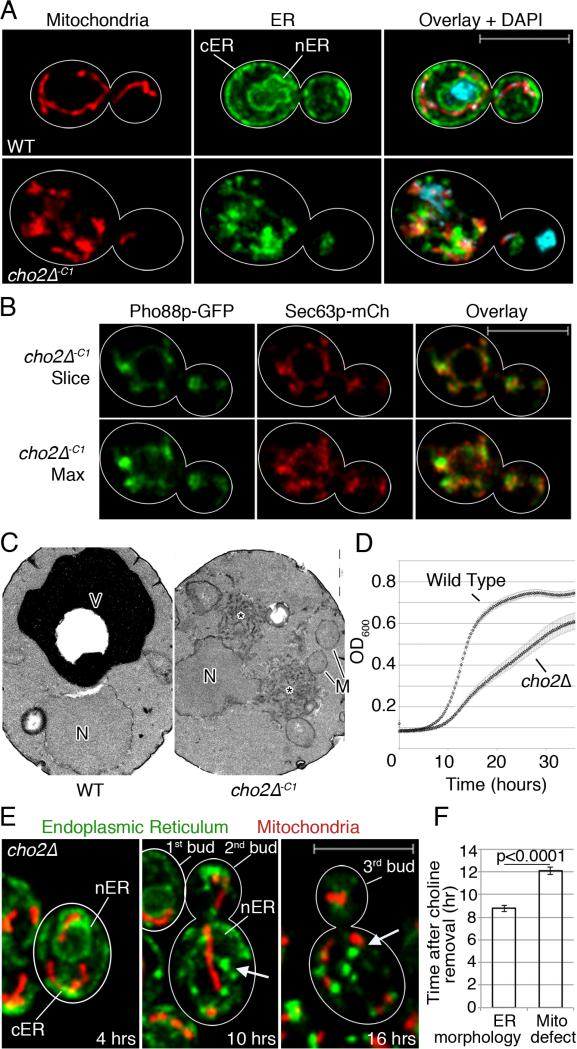
Fig. 2. Lipid imbalance triggers mitochondria and ER morphology defects.
(a) Maximum projections of of WT and cho2Δ cells expressing Cit1p-mCherry and Pho88p-GFP Cell outlines in white. Bar: 5 μm. (b) A single optical section (top) or maximum projection (bottom) of Pho88p-GFP and Sec63p-mCh in cho2Δ−c1 cells. (c) TEMs of WT and cho2Δ-c1 cells, N: nucleus. M: mitochondrion. V: vacuole. Asterisk marks abnormal membrane aggregates. Bar: 1 μm. (d) Growth of WT and cho2Δ cells. (e) Time-lapse images of cho2Δ cells perfused with choline-free media. Arrows point to abnormal ER aggregates. Bar: 5 μm (f) Mean time to organelle aggregation after choline removal +/− SEM. P-values calculated using Student's t-test. One representative trial from 3 independent trials. n>10 for each trial. See also Figure S2.
Upon acute lipid imbalance (cho2Δ-C1), there are severe defects in mitochondria and ER (Fig. 2a-c). The normal morphology of both organelles is lost: there are no tubular mitochondria or cER tubules or sheets. Polarized localization and normal interactions of mitochondria and ER with each other and at the cell periphery are also lost. nER is detectable by fluorescence and electron microscopy; however, there is almost no detectable cER, and almost no anchorage of mitochondria to the bud tip or mother cell tip (Fig. 2b-c). Instead, mitochondria and ER form abnormal fragmented structures that aggregate near each other. Ultrastructural analysis of cho2Δ−C1 cells confirm results obtained by fluorescence microscopy. We detect abnormal ER-like membrane aggregates in close proximity to the nucleus and mitochondria (Fig. 2c).
Further investigation of ER by expression of Sec63p-mCh and Pho88p-GFP in cho2Δ−C1 cells revealed that both ER proteins localize to similar structures. Interestingly, we detect subtle differences in the distribution of each fluorescently tagged protein within ER aggregates (Fig 2b). This finding provides additional evidence that lipid imbalance results in defects in ER morphology and raises the possibility that integral ER membrane proteins can segregate under these lipid stress conditions.
In wild-type cells, mitochondria and ER are continually transferred into and inherited by buds as they grow, and the amount of both organelles that are present in buds is proportional to bud size (Rafelski et al., 2012; West et al., 2011). In cho2Δ−C1 cells, aggregated mitochondria and ER are absent or present in very low levels in buds. Thus, there are also defects in the inheritance of both organelles by developing buds (Fig. 2a and 3b). We also observe a decrease in the velocity of mitochondrial movement (Fig. S2a); however, mitochondria retain mtDNA and have a more reducing matrix relative to controls (Fig. S2b-c,d). Thus, acute lipid imbalance affects the morphology, localization, motility, and inheritance of mitochondria but does not have a negative impact on maintenance of their genome or redox state.
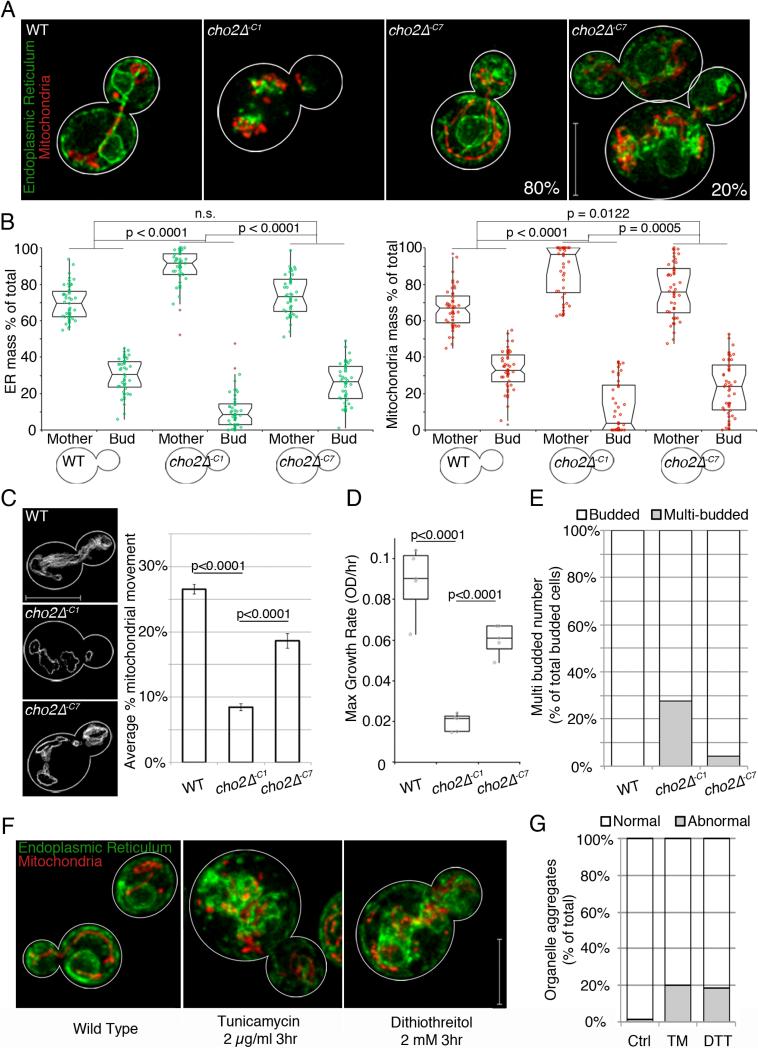
Fig. 3. Yeast cells adapt to the lipid stress associated with decreased PC biosynthesis.
(a) Maximum projections of WT and cho2Δ cells expressing Cit1p-mCherry, Pho88p-GFP. (b) Mitochondrial and ER distribution between mother cells and buds, expressed as percentage of total integrated fluorescence intensity, plotted in a notched dot box plot. n>40 for each condition. P-values from Kruskal-Wallace test with Bonferroni correction. (c) Representative traces of total mitochondrial motility (grey). Mean mitochondrial movement +/− SEM. P-values from Student's t-test, n>20 for each condition. (d) Box dot plots of maximum growth rates of indicated strains. P-values from Kruskal-Wallace test with Bonferroni correction, quintuplicate replicates for each condition. (e) Percentage of multi-budded cells (grey) as a function of total budded cells (white) in each condition, n>50 for each condition. (f) Maximum projections of mitochondria and ER after 3 hr DMSO, TM or DTT treatment. (g) Mitochondrial and ER morphology defects (grey) expressed as percentage of cells containing aggregated organelles, n>60 for each condition. For all quantitation, 1 representative trial is shown from 3 independent trials. Scale bars: 5 μm. See also Figure S3.
Previous studies revealed that severe defects in the inheritance of mitochondria or cER trigger checkpoints that inhibit cell cycle progression at cytokinesis (Babour et al., 2010; García-Rodríguez et al., 2009). Moreover, elevated ceramide levels in ER-stressed yeast result in reduced growth rates and loss of cell viability (Mousley et al., 2008). Therefore, we studied cell growth and cell cycle progression through cytokinesis in cho2Δ−C1 cells. Indeed, we observe clear defects in cell growth rate (Fig. 2d) and cytokinesis (Fig. S2d).
Next, we used long-term time-lapse imaging to assess the sensitivity of mitochondria and ER to the stress of acute lipid imbalance (Fig. 2e; Fig. S2e; Movie S1). On average, ER begins to aggregate 8 hrs after choline removal from cho2Δ cells, with ER aggregation occurring first near the nucleus and later at the cell cortex. In contrast, aggregation of mitochondria is not evident until choline has been absent from the medium for 12 hrs (Fig. 2f). Most of the mitochondrial aggregation occurs close to pre-existing ER aggregates. Thus, ER is more sensitive to the lipid imbalance associated with defects in PC biosynthesis compared to mitochondria.
Yeast cells adapt to lipid imbalance produced by defects in PC biosynthesis
Surprisingly, given the extent to which mitochondria and ER are perturbed acutely, we observe partial restoration of normal organelle morphology, localization, interactions and inheritance in cho2Δ−C7 cells. To characterize this adaptation to chronic lipid imbalance, we analyzed the morphology and inheritance of mitochondria and ER, the velocity and extent of mitochondrial movement, cell growth rates and cell cycle progression through cytokinesis. WT, WT+C, and cho2Δ+C cells have no defects in any of these parameters (Fig. S3a-d). In acutely treated cho2Δ−C1 cells, 100% of the cells analyzed exhibit severe defects in morphology of mitochondria and ER. However, with chronic lipid imbalance, we find that normal morphology of both organelles is restored in 80% of cells analyzed. In the remaining 20%, mitochondrial and ER morphology are largely similar to those observed in WT cells, except for some perinuclear aggregates of tubular mitochondria and ER (Fig. 3a).
We also find that the inheritance of both organelles and mitochondrial motility are restored to near WT levels in cho2Δ−C7 cells (Fig. 3b-c). Moreover, maximum cell growth rate reverts to 75% of that observed in WT cells (Fig. 3d) and there are no detectable defects in cytokinesis (Fig. 3e). Finally, we do not detect loss of cell viability or failure of a subset of cho2Δ cells to grow under our lipid stress conditions (data not shown). Thus, the restoration of organelle morphology and cell growth is not due to selective loss of the most severely compromised cells. Rather, these findings provide additional evidence that yeast cells adapt to lipid imbalance.
Previous studies revealed defects in ER morphology in response to ER stress (Bernales et al., 2006). Other studies revealed up-regulation of the unfolded protein response pathway (UPR), and ER associated degradation (ERAD) in cho2Δ cells with chronic lipid imbalance (Thibault et al., 2012) and in yeast and C. elegans with fatty acid unsaturation imbalance (Surma et al., 2013) (Hou et al., 2014). We find that treatment of wild-type cells with dithiothreitol (DTT) or tunicamycin (TM), two potent activators of the UPR, results in defects in mitochondria and ER that are similar those observed in cho2Δ−C7 cells: aggregation of tubular mitochondria and ER in the perinuclear region (Fig. 3f and Fig. S3e). We also observe a similarity in the penetrance of phenotype: defects in organelle morphology are evident in 20% of the DTT or TM-treated wild-type cells and in 20-30% of cho2Δ−C7 cells (Fig. 3g).
Lipid droplet biogenesis occurs at ER aggregates and is required for adaptation to lipid imbalance
TG and EE, neutral lipids in the LD core, increase during acute and chronic lipid imbalance produced by defects in PC biosynthesis (Fig. 1, Fig. S1). Therefore, we studied the effect of lipid imbalance on LDs. We found that monodansylpentane (MDH), a blue neutral lipid stain that has been used to detect LDs in mammalian cells (Yang et al., 2012), labels LDs in yeast (Fig. S4a-b). We also confirmed that wild-type cells contain 5-15 LDs per cell in close proximity to the nucleus (Fig. 4a) (Fei et al., 2008). Acute lipid imbalance does not have a major effect on LD size. However, it results in an increase in both the number and the fluorescence of MDH-stained LDs. In cho2Δ−C7 cells, there is an additional increase in both size and intensity of MDH-stained LDs compared to cho2Δ−C1 cells (Fig 4b-c). Consistent with this, quantitative analysis of yeast visualized by EM revealed that acute and chronic lipid stress results in an increase in the number of LDs per cell (see below, Fig. 5b). Thus, LDs are produced in response to acute and chronic PC biosynthetic defects.
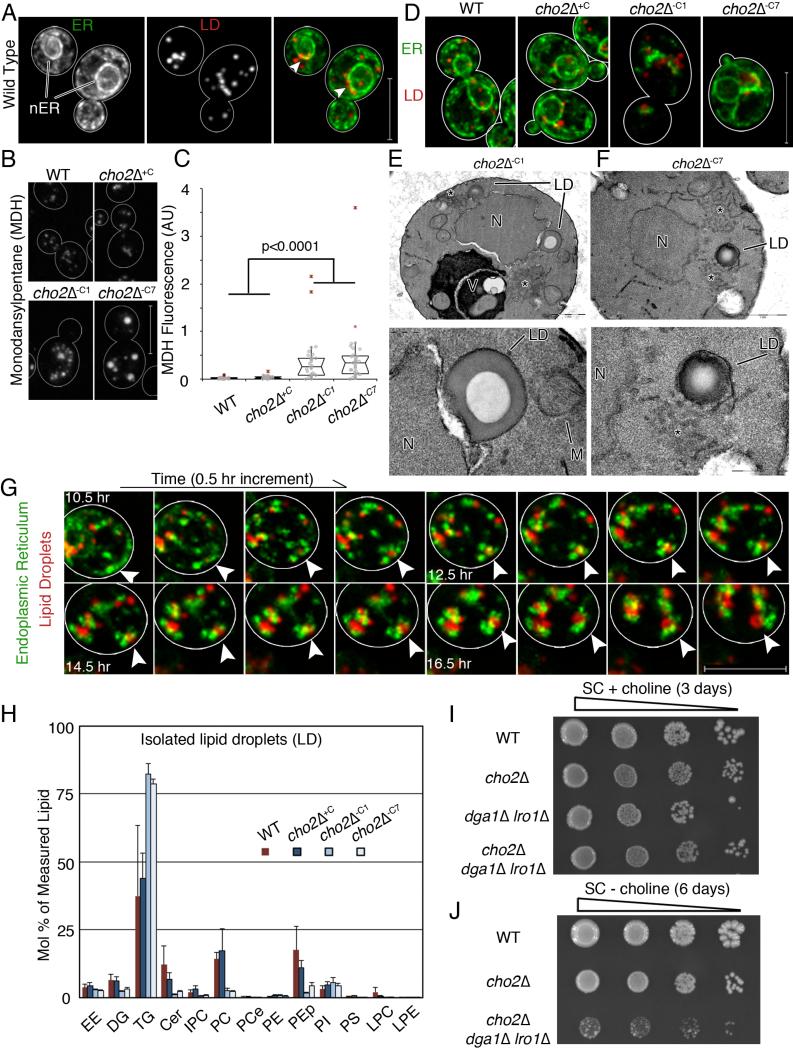
Fig 4. Lipid droplet biogenesis occurs at ER aggregates and is required for adaptation to lipid imbalance.
(a) Maximum projections of cells expressing Pho88p-GFP and Erg6p-mCherry. Arrow heads point to areas near the nucleus where LDs accumulate. Bar: 5 μm (b) Representative images of MDH-stained LDs. Bar: 5 μm. (c) Notched dot box plot of cellular MDH fluorescence. P-values from Kruskal-Wallace test with Bonferroni correction. 1 of 3 representative trials is shown, n>25 cells for each condition. (d) Representative images of ER and LDs during lipid imbalance. Bar: 5μm. (e-f) TEM of a cho2Δ cell, N: nucleus. LD: lipid droplet. V: Vacuole. LDs have a characteristic “donut” staining pattern. Asterisk marks abnormal membrane aggregates. Bottom panels show enlarged views of LDs in the respective top panels. Bar: 1 μm (d-e top), 200 nm (d bottom), 500 nm (e bottom). (g) Representative time-lapse frames of ER and LDs in a cho2Δ cell after transfer to choline-free medium. Arrow heads point to the same spot in all images. Bar: 5 μm. (h) Lipids analyzed as in Fig. 1 are represented as mean +/− SEM. (i) Dot assay of serially diluted (1:10) strains on SC + choline media. (j) Dot assay of serially diluted (1:10) strains grown to mid-log phase in choline-containing media and plated on choline-free SC media. See also Figure S4.
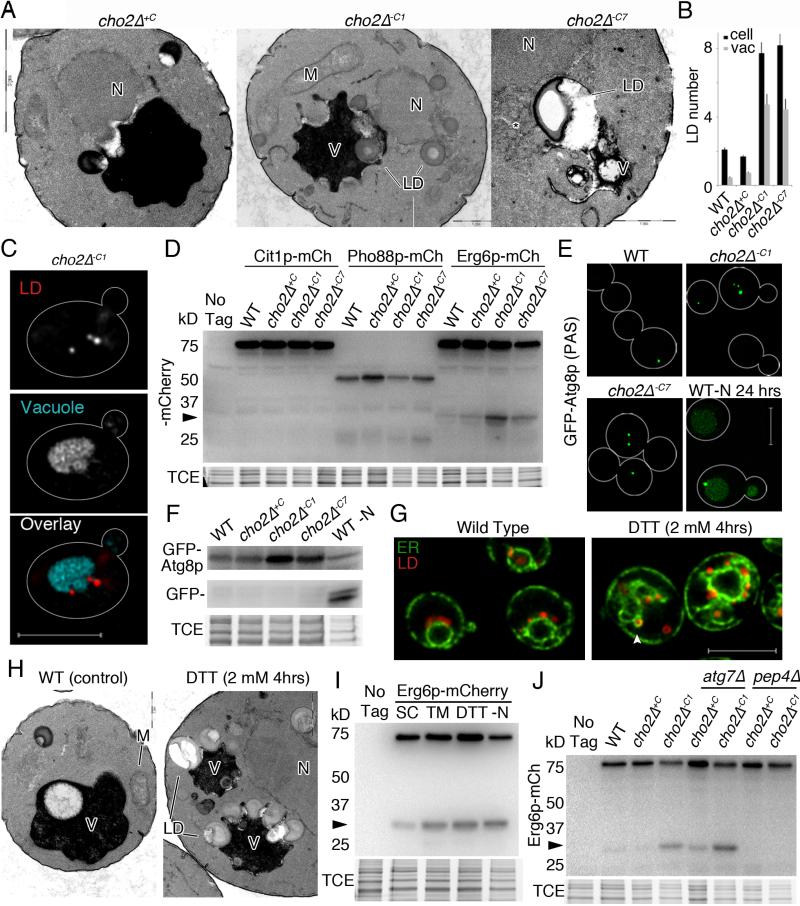
Fig. 5. Stress-induced LDs are degraded in the vacuole in a process that resembles microautophagy.
(a) TEM of cho2Δ cells showing interactions of LDs and vacuoles. N: nucleus. LD: lipid droplet. M: mitochondria. V: vacuole. Bars, from left to right: 2 μm, 1 μm, and 1 μm . (b) Quantitation of LD abundance in cho2Δ cells visualized by TEM. Mean +/− SEM. (c) Representative image of cho2Δ cells expressing Erg6p-mCh and stained with FM-464. Images are single slices from deconvolved wide-field z-series. Bar: 5 μm. (d) Representative western blot of cells expressing organelle-targeted mCherry reporters of autophagy. Arrow points to protease-resistant/free mCherry. (e) Maximum projections of GFP-Atg8p. Bar: 5 μm. (f) Representative western blots of GFP-Atg8p, with TCE as a load control. (g) Representative central slices of cells expressing Pho88p-GFP and Erg6p-mCherry. Arrowhead points to area near nucleus showing enlarged LDs. (h) TEM of untreated and DTT-treated cells. Labels are defined in (a). Bar: 500 nm. (i-j) Representative western blots showing vacuolar degradation of Erg6p-mCh to free mCherry (arrow). TCE: load control. All experiments (except TEM): 1 representative trial is shown from 3 independent trials. See also Figure S5.
Imaging of Pho88p-GFP (an ER marker) and Erg6p-mCherry (a LD marker) revealed a close relationship between LDs and the ER. In cho2Δ−C1 cells, LDs are in close proximity to ER aggregates. In cho2Δ−C7 cells, in which the majority of the ER has reverted to WT morphology and localization, residual ER aggregates that are present are often associated with large LDs (Fig. 4d and Fig. S4c). Ultrastructural analysis confirmed that LDs are frequently associated with nER, and with membrane aggregates, presumably cER (Fig. 4e-f and Fig S4d). In some cases, surfaces at the interface between LDs and membranes have different staining properties compared to those of either organelle, suggesting that there is a specialized structure at that site. Moreover, using time-lapse imaging, we obtained evidence that LDs form, grow, and persist in association with developing ER aggregates in cho2Δ cells during acute lipid imbalance (Fig. 4g; Movie S2).
As described above, PE, PG, and PI increase during acute loss of PC biosynthesis. However, during chronic loss of PC biosynthesis, when TG levels increase, the levels of these phospholipids decrease. This raises the possibility that excess lipids are converted to TG and removed from membranes as LDs. If this is the case, then TG levels should increase in LDs in yeast exposed to lipid stress and deletion of enzymes that convert phospholipids to TG should have a negative effect on yeast challenged with lipid stress. Indeed, we detect a 2-fold increase in TG in LDs isolated from cho2Δ−C1 or cho2Δ−C7 cells compared to WT or cho2Δ+C cells (Fig. 4h).
Moreover, deletions of TG biosynthetic enzymes prevent adaptation to lipid stress. Lro1p catalyzes direct esterification of diacylglycerol using the sn-2 acyl group from phospholipids to generate TG (Dahlqvist et al., 2000). Acyl groups from phospholipids can also be transferred to CoA via sequential phospholipase- and acyl-CoA synthetase-catalyzed reactions. Acyl-CoA then serves as an acyl donor for TG production by DGA1 (Oelkers et al., 2002). Deletion of DGA1 and LRO1 blocks TG biosynthesis and decreases LDs (Jacquier et al., 2011).
All strains examined (WT, cho2Δ, dga1Δ lro1Δ and cho2Δ dga1Δ lro1Δ) exhibit similar growth rates in choline-supplemented media (Fig. 4i). Removal of choline does not affect the growth rate of dga1Δ lro1Δ cells (Fig. S4e). However, the growth of cho2Δ dga1Δ lro1Δ yeast on choline-free media is severely compromised compared to cho2Δ cells (Fig. 4j). Thus, Dga1p and Lro1p are required for viability and adaptation to lipid imbalance resulting from defects in PC synthesis, presumably due to their role in conversion of excess phospholipids to TG, which are ultimately sequestered in LDs.
Stress-induced lipid droplets are degraded in the vacuole in a process that resembles microautophagy
Microautophagy is an autophagic pathway whereby cellular constituents undergo direct, autophagosome-independent uptake by the vacuole or lysosome (Li et al., 2012). Our ultrastructural and live-cell fluorescent studies revealed that LDs undergo delivery to the vacuole during lipid stress in a process resembling microautophagy (Fig. 5a, 5c). Specifically, we detect association of LDs with vacuoles, invagination of the vacuolar membrane at sites of contact with LDs, and LDs within vacuoles in cho2Δ−C1 and cho2Δ−C7 cells. We also observe changes in the appearance of membranes at the site of contact between LDs and vacuoles, indicating that a unique structure is formed at that site. Finally, quantitative analysis of cells visualized by EM revealed an increase in the number of LDs in vacuoles in cho2Δ−C1 and cho2Δ−C7 cells (Fig 5b).
To further characterize lipid stress-induced lipophagy and determine whether other organelles are targeted to the vacuole for degradation under these conditions, we tagged Cit1p, Pho88p and Erg6p, marker proteins for mitochondria, ER and LDs, respectively, with mCherry and assessed targeting of those proteins to the vacuole by analysis of the degradation of tagged proteins to free mCherry, an established assay for autophagy (Klionsky et al., 2012). We confirmed previous findings that nitrogen starvation induces degradation of the markers for mitochondria, ER and LDs (Fig. S5a). Equally important, we find that lipid stress induces degradation of the LD marker but not markers for mitochondria and ER (Fig. 5d), and that degradation of Erg6p-mCh requires a vacuolar protease, Pep4p (Fig. 5f). Thus, lipid stress induces LD-specific autophagy.
To determine whether LD degradation requires autophagosome formation, we studied Atg8p, a small membrane bound ubiquitin-like protein that localizes to pre-autophagosomes and autophagosomes and is required for efficient autophagosome formation (Kirisako et al., 1999) (Suzuki et al., 2001). We confirmed that GFP-Atg8p localizes to punctuate structures, the phagophore assembly site (PAS), in 1 out of 4 mid-log phase wild-type yeast. We also confirmed that nitrogen starvation results in autophagosome formation, targeting of Atg8p to the vacuole (Fig. 5e), and degradation of GFP-tagged Atg8p to free GFP (Fig. 5f). In addition, we find that Atg8p localization and expression is similar in WT and cho2Δ+C cells, but find a 4-fold increase in steady-state levels of GFP-Atg8p and PAS in cho2Δ−C1 and cho2Δ−C7 cells (Fig 5e-f, Fig. S5b). Equally important, we do not observe Atg8p in autophagosomes, delivery of Atg8p to the vacuole, or degradation of Atg8p-GFP to free GFP in cho2Δ−C1 or cho2Δ−C7 cells. Thus, lipid imbalance does not induce measureable amounts of macroautophagy of the markers studied or of GFP-Atg8p. Moreover, we find that lipid imbalance induces LD degradation in the vacuole, but no obvious association of LDs with autophagosomes. This is additional evidence that LD degradation occurs by a process resembling microautophagy.
Because lipid stress induces ER stress, we tested whether TM- or DTT-induced ER stress induces LD biogenesis and microlipophagy. Indeed, we find that either treatment results in an increase in LD abundance (Fig. 5g) and (Fei et al., 2009), and both, light and electron microscopy reveals that LDs induced by ER stress (DTT) are associated with perinuclear ER aggregates (Fig. 5g and Fig. S5f). Our ultrastructural studies also reveal that LDs are taken up into the vacuole by a process that resembles microautophagy during ER stress (Fig. 5h). Specifically, we find that a LD marker protein (Erg6p-mCh) is degraded with no obvious degradation of ER marker proteins in TM- or DTT-treated yeast (Fig. 5i and Fig. S5g).
Moreover, the localization of GFP-Atg8p in yeast undergoing ER stress is similar to that observed in yeast exposed to lipid stress. Specifically, DTT or TM treatment results in an increase in the number of PAS (Fig. S5c-d) and increased steady-state levels of GFP-Atg8p (Fig. S5e). However maturation of PAS structures into autophagosomes, or delivery of GFP-Atg8p to the vacuole was not detected. These findings indicate that ER stress also induces LD biogenesis and microautophagy, and raise the possibility that the LD biogenesis and microautophagy observed in cho2Δ cells undergoing acute lipid stress are due to lipid stress-induced ER stress.
Finally, we examined the effect of deletion of ATG7 in cho2Δ cells during lipid stress. Atg7p is a core autophagy protein and dual-specificity E1 ubiquitin ligase protein that is required for activating Atg8p for multiple autophagic pathways. Surprisingly, we find that lipid stress induced-LD degradation does not require ATG7 (Fig. 5j). Indeed, although LD levels are higher in cho2Δ atg7Δ double mutants relative to cho2Δ cells under chronic lipid stress (Fig. S5i-j), LD autophagy actually occurs to a greater extent in atg7Δ cho2Δ−C1 compared to cho2Δ−C1 cells (Fig. 5j). In light of these findings, we conclude that lipid stress-induced LD microautophagy is a new form of specific autophagy that we will refer to as microlipophagy.
Chaperone proteins are recruited to ER aggregates, and LD fractions are enriched in ubiquitinated proteins
Previous studies revealed that lipid stress in cho2Δ cells results in activation of the UPR and ERAD (Thibault et al., 2012). Other studies suggest a functional connection between ER stress and the cytosolic heat shock proteins (Schroder et al., 2003) (Liu and Chang, 2008). Therefore, we studied the effect of lipid stress on Hsp104p, a cytosolic oligomeric ATPase from the HSP100 family of proteins (Schirmer et al., 1996) that binds to and mediates refolding of misfolded proteins (Glover and Lindquist, 1998). Hsp104p localizes to cytosolic compartments that contain misfolded proteins including IPOD (insoluble protein deposit), JUNQ (juxtanuclear quality control compartment) (Kaganovich et al., 2008) and Q-bodies (Escusa-Toret et al., 2013).
We confirmed that Hsp104p tagged with mCherry localizes to small punctate structures in 50% of mid-log phase wild-type yeast, and that heat shock results in an increase in the number of Hsp104p-containing small punctate structures (Fig. S6a-b). In addition, we found that Hsp104p levels and distribution are similar in WT and cho2Δ+C cells (Fig 6a-b, Fig. S6c). In contrast, in cho2Δ−C1 cells, steady-state levels of Hsp104p increase (Fig. 6a), and Hsp104p localizes to large aggregates (Fig. 6b, Fig. S6c). In cho2−C7 cells, Hsp104p levels are elevated; however, yet the protein does not localize to large aggregates (Fig. 6b
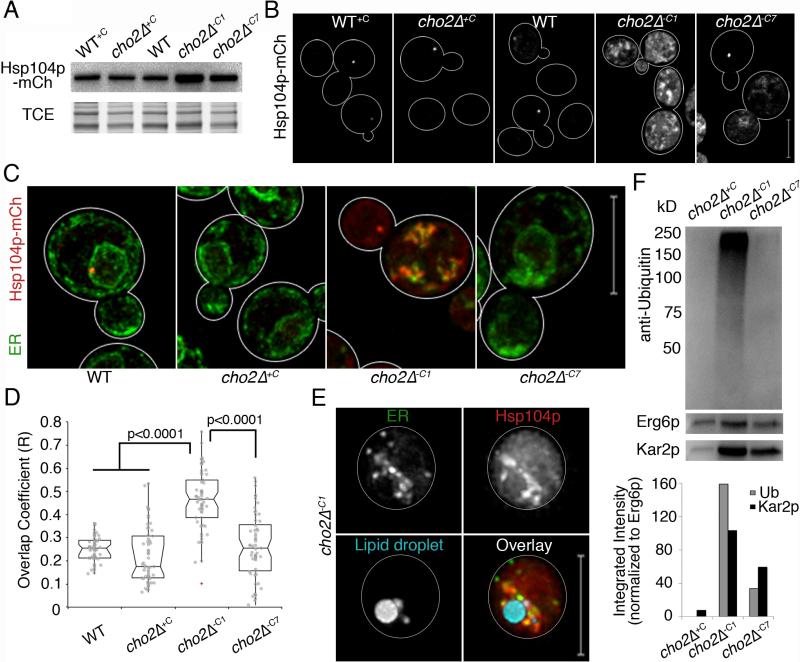
Fig. 6. Chaperone proteins are recruited to ER aggregates and LD fractions are enriched in ubiquitinated proteins.
(a) Western blot of Hsp104p-mCh. TCE: load control. (b) Maximum projections of Hsp104-mCh. (c) Representative images of Pho88p-GFP and Hsp104-mCh. (d) Notched dot box plots of colocalization between ER and protein aggregate fluorescence in (c) using Manders’ overlap coefficient (R). P-values from Kruskal-Wallace test with Bonferroni correction. n>50 for each condition. (e) Representative images of cells as in (c) with LDs labeled with MDH. Bar: 5 μm. (f) Upper panel: Representative western blot of ubiquitinated proteins in LDs isolated as in Fig. 4h. LDs isolated from cho2Δ+C cells were used as controls. Erg6p-mCh was used as a loading control. Blots were also probed for Kar2p as an internal ER chaperone. Lower panel: Quantitation of ubiquitin and Kar2p levels normalized to Erg6p levels. 1 representative trial is shown from 3 independent trials fpr all experiments. All scale bars: 5 μm. See also Figure S6.
In light of these findings, we studied the spatial relationship between ER and Hsp104p. In cho2Δ−C7 cells, Hsp104p abundance and localization resemble those observed in WT cells (Fig. 6c-d). Hsp104p localizes to 1-2 punctate, perinuclear structures per cell. In contrast, in cho2−C1 cells, Hsp104p forms large aggregates that are enriched at and co-localize with ER aggregates. Furthermore, live-cell imaging of cho2Δ−C1 cells revealed that ER aggregates are enriched with Hsp104p and are in close proximity to LDs (Fig. 6e). Thus, unfolding of ER proteins occurs during lipid imbalance produced by inhibition of PC biosynthesis, and is largely restored during chronic lipid imbalance. Moreover, since Hsp104p localizes to ER aggregates and does not localize to other membranes during acute lipid imbalance (data not shown), the primary site for protein unfolding under these conditions is the ER.
Since lipid imbalance leads to protein misfolding in the ER, and unfolded ER proteins appear to elicit LD biogenesis, we examined whether damaged ER proteins are removed from the ER by LDs. Damaged proteins are marked for degradation by polyubiquitination (Vembar and Brodsky, 2008). Moreover, Kar2p is a chaperone that binds to unfolded proteins in the ER lumen (Normington et al., 1989). We isolated LDs and ER from cho2Δ−C1 and cho2Δ+C cells and found that the protein profiles of LD and ER fractions are clearly distinct. Moreover, similar amounts of the ER marker protein Pho88p are found in LDs isolated from cho2Δ-c1 and cho2Δ+C cells, indicating that the levels of ER contamination in both fractions are similar (Fig. S6d). Interestingly, we found that Kar2p and polyubiquitinated proteins are associated with LDs isolated from cho2Δ cells during lipid imbalance, and are enriched in LDs isolated from cho2Δ−C1 cells (Fig. 6f). Furthermore, the level of polyubiquitinated proteins as a function of total protein in cho2Δ−C1 cells is 1.5 – 1.6 fold greater in LD fractions compared to ER fractions (Fig. S6d). Because LDs form at ER aggregates and contain an ER chaperone and protein unfolding occurs in ER aggregates in yeast undergoing acute lipid stress, we favor the interpretation that the observed LD-associated polyubiqutinated proteins are misfolded ER proteins.
Stress-induced microlipophagy is required for adaptation to lipid stress and is regulated by a previously uncharacterized protein
We used next-generation RNA sequencing (RNAseq) to assess changes in gene expression associated with acute lipid imbalance in cho2Δ cells. Using 1.5 log2-fold differences as a cut-off for changes in gene expression, we detect changes in the expression of hundreds of genes. We grouped gene ontology (GO) terms related to transcripts that are elevated or reduced in response to acute lipid stress using the Revigo protocol (Fig. 7a, Fig. S7a, Table S1) (Supek et al., 2011). Lipid imbalance results in up-regulation of stress processes, including chaperones linked to heat and abiotic stress; catabolism of lipids, proteins and carbohydrates; autophagy; vacuolar proteases; and carbohydrate transporter genes. Indeed, the majority of the genes that encode autophagy proteins or heat shock proteins are up-regulated (Fig. S7b-c). Groups of GO terms also clustered around oxidation-reduction related terms, mainly involving mitochondrial homeostasis. Transcripts that were down-regulated in response to acute defects in PC biosynthesis were almost exclusively related to cytosolic translation (Fig. S7a).
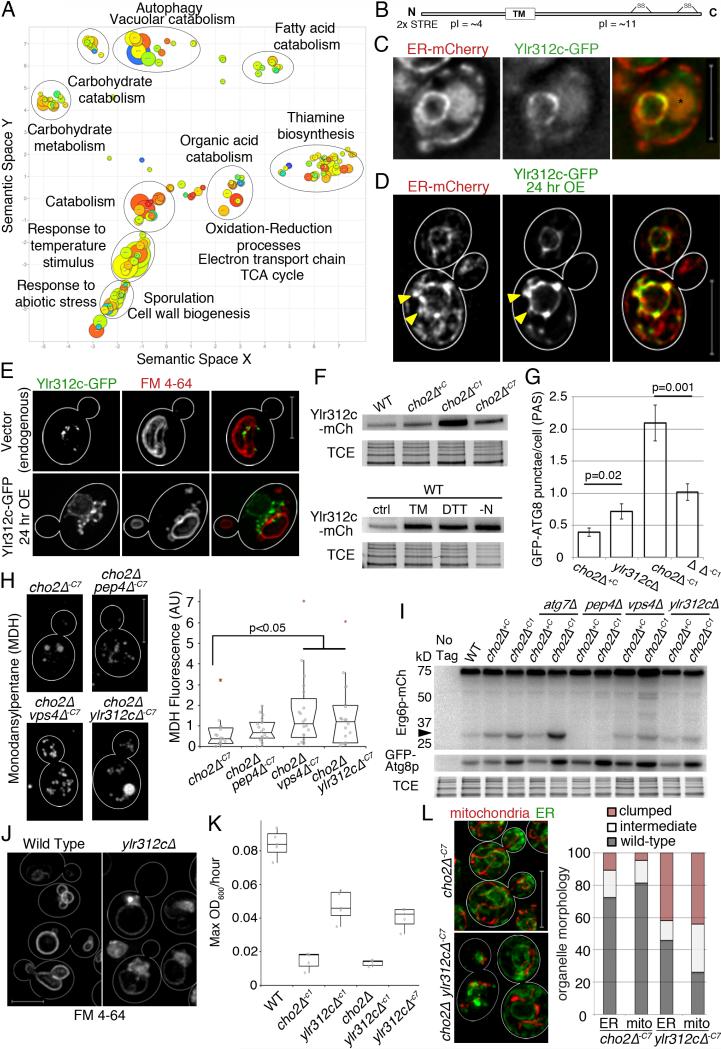
Fig. 7. Stress-induced microautophagy is regulated by the ESCRT complex and a previously uncharacterized protein.
(a) Revigo plot of up-regulated GO terms from RNAseq data identifying differentially expressed mRNAs between WT cells and cho2Δ−C1 cells. (b) Ylr312p predicted domains and properties. (c) Representative images of cells expressing Pho88p-mCh and genomic Ylr312p-GFP. Asterisk marks vacuole autofluorescence. Images are single slices from deconvolved wide-field z-series. (d) Representative images of cells expressing genomic Pho88p-mCh and a GAL construct containing Ylr312p-GFP, induced for 24 hr (24 hr OE). Maximum projections are shown of 2 adjacent slices from deconvolved wide-field z-series. Arrowheads point to Ylr312c-containing nER projections. (e) Representative images of Ylr312c-GFP expressed from the endogenous locus, and OE Ylr312p-GFP. Vacuole visualized with FM4-64. Maximum projections of deconvolved wide-field z-series are shown. (f) Representative Ylr312c-mCh western blot. TCE: load control. (g) Quantification of GFP-Atg8p puncta (PAS and autophagosomes) per cell. Mean +/− SEM. P-values from Student's t-test, n>30 for each condition. (h) Representative images of MDH stained LDs. Quantitation of MDH fluorescence as for Fig. 4c. (i) Analysis of microlipophagy was carried out as for Fig. 5j. (j) Maximum projections of cells stained with FM4-64. (k) Maximum growth rates were calculated as for Fig. 2d. (l) Left: Mitochondria and ER imaged as for Fig. 3a. Right: Quantitation of defects in organelle morphology and distribution. 1 representative trial is shown from 3 independent trials for all experiments. Scale bars: 5 μm. See also Figure S7.
Many of the genes that exhibited changes in expression could not be grouped because they were previously uncharacterized or because their GO terms were either not enriched or were not well annotated. We confirmed that genes involved in the ER-UPR are up-regulated in response to acute lipid imbalance (Thibault et al., 2012); however, since all of these genes have a number of roles or are not principally GO annotated for the UPR, they are not shown in our analysis. Other gene products like Ire1p, which relies on post-translational regulation, show dampened transcriptional regulation relative to genes whose primary regulation is through mRNA abundance. Our transcriptome analysis revealed several previously uncharacterized open reading frames (ORFs) that are up-regulated in response to lipid stress. One of these ORFs, YLR312c is up-regulated > 32-fold during acute lipid stress. This ORF contains two repeats of a stress response element (STRE) ~ 500 bp upstream of the promoter, suggesting a role in a stress response.
We tagged YLR312c at its chromosomal locus with GFP or mCherry and did not detect any obvious defect in growth rates, or in the morphology of ER or mitochondria in yeast that express tagged YLR312c. Live cell imaging of yeast expressing GFP-tagged YLR312c revealed that the protein localizes to ER. Specifically, it is enriched on nER and detectable in a limited number of cER structures (Fig. 7c). YLR312c protein contains a putative transmembrane domain, an amino terminal region that has an isoelectric point (pI) of 4, and a C-terminal region that has a pI of 11 (Fig. 7b). Thus, it is likely that YLR312c encodes an integral nER protein whose amino terminal region is predicted to face the cytosol and the C-terminal region is predicted to be in the ER lumen (Kall et al., 2004). Interestingly, when tagged YLR312c is overexpressed, it localizes to protrusions that extend from the nER and accumulates at sites of close contact between nER and the vacuole (Fig. 7d-e; Fig. S7d).
We also obtained evidence for a link between YLR312c and lipid stress-induced microlipophagy. Specifically, we find that the steady-state level of YLR312c protein increases during lipid stress,TM- or DTT-induced ER stress, and nitrogen starvation (Fig. 7f). Formation of Atg8p-containing PAS does not occur in cho2Δ ylr312cΔ−C1 cells or in ylr312cΔ cells treated with DTT or TM. However autophagosomes form and are delivered to the vacuole in ylr312cΔ cells during nitrogen starvation (Fig. 7g; Fig. S7g-h). This suggests that PAS formed during ER stress are different from the PAS formed during starvation and support a role for YLR312c in ER stress-induced PAS formation.
Moreover, deletion of YLR312c results in defects in lipid stress-induced microlipophagy. Specifically, the levels of MDH-stained LDs are higher in cho2Δ ylr312cΔ−C7 cells compared to cho2Δ−C7 cells (Fig. 7h). Moreover, we do not detect degradation of Erg6p-mCh to free mCh in cho2Δ ylr312cΔ−C1 or cho2Δ ylr312c−C7 cells (Fig. 7l). Thus, YLR312c is up-regulated by lipid stress and required for lipid stress-induced microlipophagy. In light of these observations, we refer to the YLR312c gene as ESM1 for ER stress induced microlipophagy gene 1.
Our studies also revealed a role for endosomal sorting complexes required for transport (ESCRT) in Esm1p-mediated microlipophagy. The ESCRT machinery has diverse roles, from membrane bending and scission for endocytic membrane transport and multivesicular body (MVB) formation, to cytokinesis and viral budding (Schuh and Audhya, 2014). Recent studies revealed that ESCRT mediates selective degradation of poly-ubiquitinated vacuole proteins. During this process, poly-ubiquitinated vacuolar membrane proteins undergo MVB-dependent, ATG-gene independent targeting to the vacuole for degradation (Li et al., 2015).
We find that esm1Δ mutants display a class E vacuole morphology defect that is characteristic of ESCRT dysfunction: accumulation of an enlarged MVB adjacent to the vacuole (Fig. 7j) (Raymond et al., 1992). To determine if ESCRT is required for lipid stress-induced microlipophagy, we studied the effect of deletion of VPS4, which catalyzes the final step in ESCRT maturation, in this process. We do not detect degradation of Erg6p-mCh to free mCh in vps4Δ cho2Δ−C1 cells (Fig. 7i). Thus, our studies support a role for the ESCRT machinery and a newly identified gene ESM1 (YLR312c) that regulates the class E compartment in yeast in stress induced microlipophagy.
Finally, we tested whether microlipophagy is required for adaptation to lipid imbalance produced by inhibition of PC biosynthesis. To do so, we studied growth rates and organelle morphology in esm1Δ yeast exposed to chronic lipid imbalance. First, we find that deletion of ESM1 results in a decrease in the rate of cell growth in wild-type yeast. Thus, Esm1p contributes to cellular function under non-stressed conditions. Equally important, the growth rate of esm1Δ cho2Δ−C7 cells is severely compromised and similar to that of cho2Δ−C1 cells (Fig. 7k). Consistent with this, we find that ESM1 is required for restoration of the morphology and distribution of mitochondria and ER in cho2Δ cells challenged with chronic lipid stress (Fig. 7l). In cho2Δ−C7 cells mitochondria and ER morphology are similar to those observed in wild-type cells and only 15-20% of the cells examined contain aggregates of mitochondria or ER. In contrast, in esm1 cho2Δ−C7 cells, >40% of the cells examined have organelle aggregates. Thus, we find that microlipophagy is required for adaption to the lipid imbalance produced by inhibition of PC biosynthesis.
Discussion
Here, we describe profound defects in yeast growth and membrane organization in response to acute lipid imbalance resulting from defects in PC biosynthesis. We also identify adaptation mechanisms to overcome effects of this glycerophospholipid imbalance, which rely on LD biogenesis and autophagy for removal of excess phospholipids and damaged proteins from the ER.
Role for LD biogenesis in adaptation to lipid stress
Our findings support the model that LDs that are produced during acute and chronic lipid stress contribute to adaptation to lipid imbalance by removal of excess phospholipids in the form of TG. We find that Lro1p and Dga1p, two enzymes that mediate conversion of phospholipids to TG and are required for LD biogenesis, are required for adaptation to lipid imbalance in cho2Δ yeast. Consistent with this, there is an increase in LDs, in TG species within cells, and in TG within LDs in cho2Δ−C1 cells. Thus, our studies support the model that excess phospholipids are converted to TG, which are sequestered in LDs as cells adapt to lipid stress produced by defects in PC biosynthesis.
We also identified defects in morphology, distribution, and inheritance of ER and mitochondria during acute lipid imbalance. Moreover, we find that ER is more sensitive to lipid imbalance compared to other organelles. Indeed, it is likely that ER is the primary site of dysfunction, as a number of stress response pathways converge on the ER during lipid imbalance.
In cells adapted to chronic lipid imbalance, PE, PA, PS, PI, and Cer are partially or completely restored to wild-type levels, so the restoration of the morphology and localization of ER and mitochondria may arise from the restoration of any or all of these phospholipids. However, PE is an abundant, conical phospholipid that assembles into reversed nonlamellar structures and can generate negative curvature and membrane stress (Gruner, 1985). Moreover, shortening and increasing the saturation of acyl side chains of PE reduces negative curvature membrane stress. Therefore, we propose that decreasing PE is a key step in adaptation. Reduction in PE levels would reduce negative membrane curvature, contributing to restoration of ER and mitochondrial organization, and, in turn, cell growth.
Role for LDs in ER Quality Control in Yeast Challenged with Lipid Stress
Previous studies revealed a role for ER-associated protein degradation (ERAD) in identifying unfolded proteins in the ER and retrotranslocating them to the ER surface, where they are ubiquitinated and targeted for degradation by the proteasome (Brodsky, 2012). Other studies in mammalian cells indicate that misfolded GPI-anchored proteins are not readily degraded by ERAD and are eliminated from the ER by ERAD-independent vesicular transport followed by degradation in the lysosome (Ashok and Hegde, 2009; Satpute-Krishnan et al., 2014). Thus, subsets of unfolded proteins with distinct physical properties can be removed from the ER and degraded by ERAD-independent pathways. Indeed, it has been proposed (Ploegh, 2007) that LDs may serve as “escape hatches” for removal of damaged ER proteins from the organelle. However, there has been little evidence for this model.
Here, we show that during phospholipid imbalance and ER stress, LDs increase in number and are associated with polyubiquitinated proteins and the ER chaperone Kar2p. Importantly another ER marker protein Pho88p, a protein with no known chaperone or protein folding function, is not enriched in LDs under these conditions. Overall, we find that ER stress occurs during lipid stress, LDs form at ER aggregates under these conditions, and polyubiquitinated proteins and Kar2p are enriched in LDs isolated from yeast under acute lipid stress. Therefore, it is possible that proteins that are unfolded as a result of lipid imbalance in ER are removed from the organelle by LDs that form and bud from the ER. This model is consistent with the escape hatch model for ER quality control.
Role for Esm1p and microlipophagy in adaption to lipid stress
Selective autophagy, removal of specific cellular constituents such as mitochondria, ER, peroxisomes, and ribosomes, can occur by macroautophagy, microautophagy or in mammalian cells by chaperone-mediated autophagy. During macroautophagy, cellular constituents are encapsulated by a double membrane structure, the autophagosome, and delivered to the vacuole. This process requires core autophagy genes ATG1-10, ATG12-14, ATG16 and ATG18 (Feng et al., 2015). Microautophagy, the process whereby organelles are taken up into the vacuole by direct interaction with the vacuole and invagination of the vacuolar membrane, is less well characterized. In yeast, mitochondria, ER, LDs, and parts of the nucleus can be degraded by microautophagy. Core ATG genes are required for all forms of selective microautophagy in yeast except ER microautophagy (Kissova et al., 2007; Kvam and Goldfarb, 2007; Schuck et al., 2014; van Zutphen et al., 2014).
We find that that degradation of LDs that form in response to lipid stress requires Pep4p, a vacuolar protease, but does not require autophagosome formation or a core ATG gene (ATG7). Instead, it occurs by direct interaction of LDs with the vacuole, invagination of the vacuolar membrane and budding of LD into the vacuole. Indeed, sites of interaction between LDs and the vacuole membrane exhibit distinct staining properties, suggesting that a specific structure forms at that site.
LD microautophagy described in this study is distinct from that induced by starvation. First, the processes occur under different conditions: lipid and ER stress versus nitrogen starvation. Second, starvation-induced LD microautophagy requires core ATG genes including ATG7 and ATG8, but not ATG11 or ATG20 (van Zutphen et al., 2014). In contrast, lipid stress-induced microlipophagy does not require ATG7.
Our studies also reveal that this stress-induced microlipophagy is regulated by a newly identified stress response factor, which we refer to as ER stress-induced microlipophagy protein 1 (Esm1p). ESM1 (YLR312c) contains two stress response elements 500 bp upstream from its promoter and has negative genetic interactions with two proline disulfide isomerases (PDI1 and MPH2) that mediate oxidative protein folding in the ER lumen (Uetz et al., 2000) (Kim et al., 2009). Indeed, deletion of ESM1 and PDI1 is lethal in yeast. We find that ESM1 is required for lipid stress-induced microlipophagy, and identify a role for the ESCRT complex in that process. Specifically, esm1Δ cells exhibit a class E vacuolar morphology defect characteristic of loss of ESCRT function. Moreover, deletion of VPS4 and the associated defects in ESCRT assembly result in failure to induce microlipophagy in yeast undergoing acute lipid stress. Finally, we find that microlipophagy is required for adaptation to lipid stress.
Collectively, we find that LD biogenesis and microautophagy are required for adaptation of lipid and ER stress produced by inhibition of PC biosynthesis in yeast. LDs produced by these stressors remove excess phospholipids and potentially unfolded proteins from the ER. LD autophagy induced by lipid and ER stress 1) is distinct from that induced by starvation, 2) relies on Esm1p, a previously uncharacterized nER protein, and ESCRT function, and 3) may contribute to adaptation to lipid and ER stress by removing damaged ER proteins. During the preparation of this manuscript, another group reported that YLR312c has the capacity to bind to Atg8p, is required for autophagy of the nER in response to rapamycin treatment and promotes yeast cell survival upon long-term exposure to nitrogen starvation (Mochida et al., 2015). They refer to the gene as ATG39. Thus, it is possible that ESM1/ATG39 has broader roles in protein homeostasis beyond that which occurs in response to lipid and ER stress produced by inhibition of PC biosynthesis.
Materials and Methods
Assays
Protocols describing lipidome analysis, RNAseq, western blotting, electron microscopy, yeast strains and primers, growth conditions, fluorescence microscopy, isolation of LD and ER by subcellular fractionation and detailed image quantitation can be found in the supplemental information.
Long-term time-lapse imaging
For extended time-lapse imaging (>15 min), cells were loaded into a CellASIC (EMD Millipore, Billerica, MA) Y04 imaging plate. All images and image series were imported into Volocity libraries (Perkin Elmer, Waltham, MA) for deconvolution and quantitation.
Image analysis
Organelle aggregation
During normal cell cycles, mitochondria and ER do not aggregate in cytosolic clusters. The earliest time that organelle aggregation occurred after removal of choline from the media in WT and cho2Δ strains was determined by visual inspection of time-lapse images of cells expressing organelle-targeted fluorescent proteins.
Organelle distribution
Distribution of mitochondria and ER was calculated as integrated fluorescence intensity in the mother or bud relative to integrated intensity of the entire cell.
Total mitochondrial motility
Mitochondrial motility was assayed as previously described (De Vos and Sheetz, 2007). Mitochondria were imaged using Cit1p-mCherry, collected as 6 μm Z-stacks with 1 μm Z-spacing, and acquired every 30 sec for 7.5 min. To calculate the percentage of mitochondrial motility, the number of pixels that changed position was divided by the total number of pixels.
Mitochondrial redox status
Mitochondrial redox status was assayed as previously described (Vevea et al., 2013).
Mitochondrial velocity
Z-series of cells containing Cit1p-mCherry were collected with 1-μm spacing over a depth of 6 μm, every 1 sec for 1 min.
Multibudded cell analysis
Multibudded cells were defined as cells with 2 or more buds.
UPR organelle aggregation assay
Aggregates of ER and mitochondria were defined as organelle clumps, typically in the vicinity of the nucleus. Numbers of affected cells were compared to total number of cells (single and budding cells).
Analysis of GFP-Atg8p-labeled PAS
Budding cells were analyzed for punctate or circular structures representing PAS and autophagosomes respectively. These structures were manually counted on a cell-by-cell basis in Volocity and recorded in Microsoft Excel (Redmond, WA).
Statistical methods and data representation
All data were evaluated for normal distribution using the Anderson Darling test for normality. Based upon this, P-values were determined using the Kruskal-Wallace test with pairwise comparisons and Bonferroni correction or with a two-tailed Student's t-test assuming unequal variance. The Analyze-it (Leeds, UK) plugin for Microsoft Excel (Seattle, WA) was used for all statistical calculations and graph creation. Graphs are bar graphs representing the mean and SEM (standard error of the mean), or outlier notched box plots representing the median, quartiles, and 95% confidence interval, along with individual observations overlaid as dots (hollow circles).
Area under the peak was normalized to internal standard area to calculate molar lipid concentrations, which were then normalized to total across all species for each sample, with final data are presented as mean mol %. All lipid species and subclasses analyzed were found to have equal variance (data not shown) then analyzed with two-way ANOVA followed by a post hoc Dunnett's test. In all cases, *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
Supplementary Material
Acknowledgments
This work was supported by awards from HHMI 56006760 to JDV, the National Institutes of Health (NIH) 5 T32 GM007367 to EJG, NIH R01 NS056049 to GDP, from NIH-NCRR, 1S10RR023454-01 to JMM and from the Ellison Medical Foundation (AG-SS-2465) and the NIH (GM45735, GM45735S1 and GM096445) to LAP. GM45735S1 was issued from the NIH under the American Recovery and Reinvestment Act of 2009. The microscopes used for these studies were supported in part through a NIH / NCI grant (5 P30 CA13696) and obtained using funds from the NIH-NCRR (1S10OD014584) to LAP.
Abbreviations
- cER
Cortical ER
- DTT
Dithiothreitol
- ER
Endoplasmic Reticulum
- EE
Ergosteryl ester
- ESCRT
Endosomal Sorting Complex Required for Transport
- LD
Lipid Droplet
- mCh
mCherry
- MDH
Monodansylpentane
- nER
Nuclear ER
- PA
Phosphatidic acid
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- PG
Phosphatidylglycerol
- PI
Phosphatidylinositol
- PS
Phosphatidylserine
- PM
Plasma membrane
- PAS
Preautophagosomal structure
- TG
Triacylglycerol
- TM
Tunicamycin
- UPR
Unfolded Protein Response
- WT
Wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashok A, Hegde RS. Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog. 2009;5:e1000479. doi: 10.1371/journal.ppat.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babour A, Bicknell A.a., Tourtellotte J, Niwa M. A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell. 2010;142:256–269. doi: 10.1016/j.cell.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS biology. 2006;4:e423–e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Henry S.a. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Progress in lipid research. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science (New York, NY) 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Sheetz MP. Visualization and quantification of mitochondrial dynamics in living animal cells. Methods in cell biology. 2007;80:627–682. doi: 10.1016/S0091-679X(06)80030-0. [DOI] [PubMed] [Google Scholar]
- Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. The Journal of cell biology. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Wang H, Fu X, Bielby C, Yang H. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. The Biochemical journal. 2009;424:61–67. doi: 10.1042/BJ20090785. [DOI] [PubMed] [Google Scholar]
- Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015;25:354–363. doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaigg B, Simbeni R, Hrastnik C, Paltauf F, Daum G. Characterization of a microsomal subfraction associated with. Yeast. 1995;1234:214–220. doi: 10.1016/0005-2736(94)00287-y. [DOI] [PubMed] [Google Scholar]
- García-Rodríguez LJ, Crider DG, Gay AC, Salanueva IJ, Boldogh IR, Pon L.a. Mitochondrial inheritance is required for MEN-regulated cytokinesis in budding yeast. Current biology : CB. 2009;19:1730–1735. doi: 10.1016/j.cub.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini F, Smith TK. The Kennedy pathway--De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB life. 2010;62:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Gruner SM. Intrinsic curvature hypothesis for biomembrane lipid composition: a role for nonbilayer lipids. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. The Journal of cell biology. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou NS, Gutschmidt A, Choi DY, Pather K, Shi X, Watts JL, Hoppe T, Taubert S. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. Journal of cell science. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. Journal of molecular biology. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kanipes MI, Henry S.a. The phospholipid methyltransferases in yeast. Biochimica et biophysica acta. 1997;1348:134–141. doi: 10.1016/s0005-2760(97)00121-5. [DOI] [PubMed] [Google Scholar]
- Kim JH, Zhao Y, Pan X, He X, Gilbert HF. The unfolded protein response is necessary but not sufficient to compensate for defects in disulfide isomerization. The Journal of biological chemistry. 2009;284:10400–10408. doi: 10.1074/jbc.M900377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. The Journal of cell biology. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. doi: 10.4161/auto.3586. [DOI] [PubMed] [Google Scholar]
- Lamari F, Mochel F, Sedel F, Saudubray JM. Disorders of phospholipids, sphingolipids and fatty acids biosynthesis: toward a new category of inherited metabolic diseases. Journal of inherited metabolic disease. 2013;36:411–425. doi: 10.1007/s10545-012-9509-7. [DOI] [PubMed] [Google Scholar]
- Li M, Rong Y, Chuang YS, Peng D, Emr SD. Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Molecular cell. 2015;57:467–478. doi: 10.1016/j.molcel.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cellular and molecular life sciences. CMLS. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chang A. Heat shock response relieves ER stress. The EMBO journal. 2008;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn J.a., Emr SD. ER-to-Plasma Membrane Tethering Proteins Regulate Cell Signaling and ER Morphology. Developmental cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi S, Ohkuma A, Talim B, Karahashi M, Koumura T, Aoyama C, Kurihara M, Quinlivan R, Sewry C, Mitsuhashi H, et al. A congenital muscular dystrophy with mitochondrial structural abnormalities caused by defective de novo phosphatidylcholine biosynthesis. American journal of human genetics. 2011;88:845–851. doi: 10.1016/j.ajhg.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- Mousley CJ, Tyeryar K, Ile KE. Trans-Golgi network and endosome dynamics connect ceramide homeostasis with regulation of the unfolded protein response and TOR signaling in yeast. Molecular biology of .. 2008;19:4785–4803. doi: 10.1091/mbc.E08-04-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. The Journal of biological chemistry. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- Rafelski SM, Viana MP, Zhang Y, Chan YH, Thorn KS, Yam P, Fung JC, Li H, Costa Lda F, Marshall WF. Mitochondrial network size scaling in budding yeast. Science. 2012;338:822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinol AE, Cui Z, Chen MH, Vance JE. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent. Journal of Biological Chemistry. 1994;269:27494–27502. [PubMed] [Google Scholar]
- Satpute-Krishnan P, Ajinkya M, Bhat S, Itakura E, Hegde RS, Lippincott-Schwartz J. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell. 2014;158:522–533. doi: 10.1016/j.cell.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends in biochemical sciences. 1996;21:289–296. [PubMed] [Google Scholar]
- Schroder M, Clark R, Kaufman RJ. IRE1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Molecular microbiology. 2003;49:591–606. doi: 10.1046/j.1365-2958.2003.03585.x. [DOI] [PubMed] [Google Scholar]
- Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci. 2014;127:4078–4088. doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AL, Audhya A. The ESCRT machinery: from the plasma membrane to endosomes and back again. Critical reviews in biochemistry and molecular biology. 2014;49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one. 2011;6:e21800–e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surma M.a., Klose C, Peng D, Shales M, Mrejen C, Stefanko A, Braberg H, Gordon DE, Vorkel D, Ejsing CS, et al. A Lipid E-MAP Identifies Ubonvox2 as a Criticalsyss Regulatdddor of Lipid Saturation and Lipid Bilayer Stress. Molecular cell. 2013;51:519–530. doi: 10.1016/j.molcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. The EMBO journal. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne TC, Zhou C, Boldogh IR, Charalel JK, McFaline-Figueroa JR, Thoms S, Yang C, Leung G, McInnes J, Erdmann R, et al. Role for cER and Mmr1p in Anchorage of Mitochondria at Sites of Polarized Surface Growth in Budding Yeast. Current biology : CB. 2011:1994–1999. doi: 10.1016/j.cub.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink N, van der Sanden MHM, Houweling M, Helms JB, Vaandrager AB. Depletion of phosphatidylcholine affects endoplasmic reticulum morphology and protein traffic at the Golgi complex. Journal of lipid research. 2009;50:2182–2192. doi: 10.1194/jlr.M800660-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault G, Shui G, Kim W, McAlister GC, Ismail N, Gygi SP, Wenk MR, Ng DTW. The Membrane Stress Response Buffers Lethal Effects of Lipid Disequilibrium by Reprogramming the Protein Homeostasis Network. Molecular cell. 2012;48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield T.a., Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Molecular biology of the cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE, Steenbergen R. Metabolism and functions of phosphatidylserine. Progress in lipid research. 2005;44:207–234. doi: 10.1016/j.plipres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea JD, Wolken DM, Swayne TC, White AB, Pon LA. Ratiometric biosensors that measure mitochondrial redox state and ATP in living yeast cells. Journal of visualized experiments : JoVE. 2013 doi: 10.3791/50633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker DR. Phosphatidylserine decarboxylase. Biochimica et biophysica acta. 1997;1348:236–244. doi: 10.1016/s0005-2760(97)00101-x. [DOI] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV. The life of lipid droplets. Biochimica et biophysica acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Zurek N, Hoenger a., Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. The Journal of cell biology. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H-J, Hsu C-L, Yang J-Y, Yang WY. Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PloS one. 2012;7:e32693–e32693. doi: 10.1371/journal.pone.0032693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.