Fig. 2.
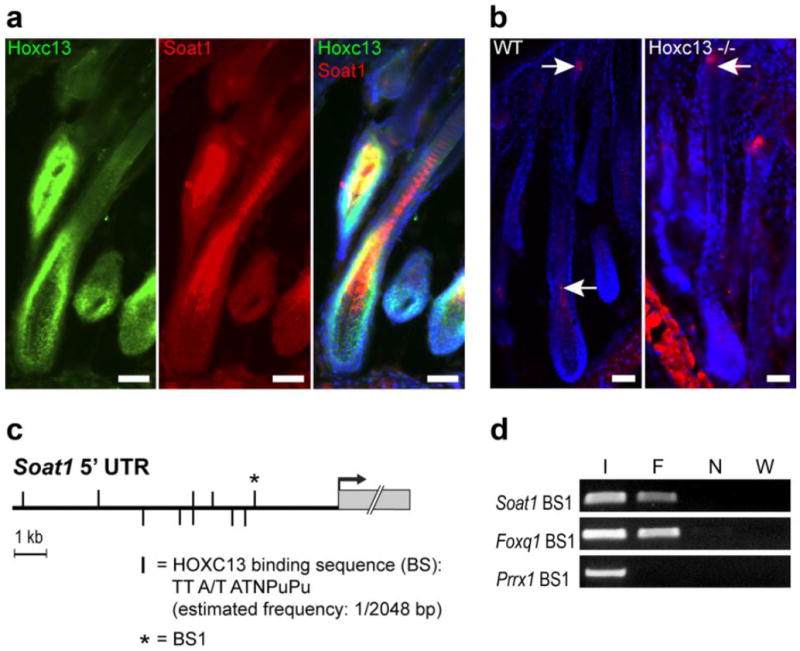
Evidence for Soat1 regulation by HOXC13 in hair. (a) Co-localization (yellow) of HOXC13 (Green) and SOAT1 protein expression (Red) by IFA in the medulla-forming compartment. (b) SOAT1 (red) expression in hair follicle bulb and sebaceous gland of wild type (WT) B6(Cg)-Tyrc-2J/J mice as indicated by arrows; SOAT1 expression is lost in the pre-medulla of Hoxc13 null (Hoxc13-/-) mice but remains in the sebaceous gland (arrow); scale bars: 50 μm. (c) Map of putative HOXC13 binding sites (vertical bars) within the 10 kb of the Soat1 5’ non-transcribed region (5’ UTR) upstream of the transcriptional start (arrow); the most proximal binding sequence (BS1) that has been subjected to ChIP analysis is marked by a star. (d) PCR analysis of HOXC13-immunoprecipitated chromatin containing Soat1 BS1 (F) and unprecipitated input DNA as positive control (I) yielded the predicted amplification products whereas ChIP DNA from untransfected cells (N) and water (W) used in negative control reactions did not. Using the same batch of immunoprecipitated DNA and a primer set specific for sequences containing a previously identified HOXC13 binding site located in the Foxq1 promoter region (Foxq1 BS1) served as a further positive control, whereas control reactions using primer sets specific for genomic regions of Prrx1 that contain no putative HOXC13binding sites failed to yield PCR products.
