Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive malignant tumors, with its 5-year survival rate lower than 5%. MicroRNAs (miR) have been known as important regulators for the tumorigenesis, progression, invasion and metastasis of various cancers. MiR-184 was found to be abnormally expressed in various cancers including glioma and oral carcinoma. The expression and functional role of miR-184 in PDAC, however, remains unclear. PDAC cell line PANC-1 was transfected with miR-184 inhibitor. Real-time PCR was used to detect the expression of miR-184 in untreated PANC-1, miR-184 inhibitor transfected PANC-1 and controlled normal pancreatic ductal epithelial cell line HPDE6c7. MTT assay was used to detect the effect of miR-184 on the proliferation of PANC-1 cells, while invasion assay and Western blotting were employed to describe the effect on cell invasion ability and expression of caspase-3, respectively. In PANC-1 cells, miR-184 was abundantly expressed. The transfection of inhibitor effectively suppressed the expression of miR-184, and further inhibited both cell proliferation and invasion abilities, in addition to the up-regulation of pro-apoptotic protein caspase 3 expression. The up-regulation of miR-184 in PDAC may facilitate the proliferation and invasion ability, and inhibit apoptosis of tumor cells, thus potentiating the occurrence and development of PDAC. MiR-184, therefore, is a potential molecular target for therapy.
Keywords: Pancreatic ductal adenocarcinoma, micro RNA inhibitor, tumor proliferation and metastasis
Introduction
With the elevating life level and changing of diet structure, the incidence and mortality of pancreatic carcinoma (PC) is increasing in recent years, making it the fourth leading malignant tumor worldwide [1]. Due to its insidious onset, PC is relatively difficult to obtain an early diagnosis. Such high-rate of misdiagnosis plus its inherent rapid progression, high degree of malignancy and difficulty in treatment, cause the 5-year overall survival rate of PC to be less than 1%, making it one of tumors with most unfavorable prognosis [2,3]. Pancreatic ductal adenocarcinoma (PDAC), which derives from epithelial cells of pancreatic duct, is the most common type of PC and occupies more than 90% of all pancreatic exocrine tumors [4,5]. In clinical practice, most PDAC patients already have tumor metastasis or invasion, thus making surgical resection impossible. Meanwhile, the insensitivity of PDAC against chemo- or radiation therapy further aggravates both life quality and survival rate of patients [6]. Therefore, it has become a research hot spot to improve the diagnostic efficiency and to develop target medication, both of which require further illustration of molecular mechanisms underlying PDAC pathogenesis.
The genetic factor underlying oncogenesis has been widely accepted, as the occurrence of cancer is a long-term aggregation of gene mutants across different locus under the direction of both inheritance and environmental factors [7]. MicroRNA, also named miRNA, small molecule RNA or mini RNA, is a family of small molecule non-coding RNA with regulatory functions and is widely existed in both animal and plant cells [8]. MiRNA can regulate the mRNA degradation and protein translation via its base-paring on target genes or inhibition on the expression of downstream target proteins [9]. MiRNA has also been suggested to be related with tumor’s occurrence and progression, as the up-regulation of some miRNAs may promote the proliferation and metastasis of tumors, while the down-regulation of some other miRNAs cause loss-of-function mutation on the tumor suppression function [10,11]. Previous studies showed the abnormal expression of mRNAs in osteosarcoma, supporting their roles as oncogenic or tumor suppressor factors. MiR-184 was recently found to have abnormal expression and participate in various processes including tumor cell growth, differentiation, invasion and metastasis [12-14]. The function of miR-184 in PDAC, however, remained unknown. This study thus investigated the expression and related regulatory mechanism of miR-184 in PDAC cells, in an attempt to provide a novel biomarker and drug target for PDAC.
Materials and methods
Cell culture
Frozen PANC-1 and HPDE6c7 cell lines (ATCC cell bank, US) were thawed in 37°C water bath can centrifuged at 1000 rpm for 3 min. Cells were re-suspended in 1 mL DMEM medium (Hyclone, US) and were cultured at 37°C with 5% CO2 in a humidified chamber. After 24 hours, cells were seeded at 1×107 per cm2 density, using high-glucose DEME medium (containing 100 U/mL penicillin and 100 μg/mL streptomycin, Hyclone, US). Cells were passed every 2~3 days, leaving log-phased PANC-1 cells as the treatment group (including untreated PDAC cell and miR inhibitor group) and HPDE6c7 cells as the control group.
MiR-184 inhibitor transfection
Oligonucleotide sequence of miR-184 inhibitor (5’-GAUCGGAGGUGCAUUCUA-3’, synthesized by Gimma Biotech, China) was used to transfect PANC-1 cells. In brief, both inhibitor expressing vector and scramble RNA control vector were mixed in 0.2 mL serum-free DMEM medium, followed by 15-min incubation at room temperature. Lipo 2000 reagents (Invitrogen, US) were then added for further 30-min incubation. Cells with confluence at 70~80% were collected, rinsed and re-suspended into 1.6 mL serum-free DMEM medium. After 6-hour incubation, normal medium with serum was added for further incubation.
Real-time PCR for miR-184 expression
Trizol reagents (Invitrogen, US) were used to extract mRNA from PANC-1 and control cells. After purification and quantification, total mRNA was used as the template to synthesize cDNA using in vitro reverse transcription kit (Invitrogen, US) and specific primers for miR-184 (Forward, 5’-TACGA CTATG TGACC TGCCT G-3’; Reverse, 5’-TGGTT CAACT CTCCT TTCCA-3) or GAPDH internal reference gene (Forward, 5’-ATCTG GAGTT TACCG CTGG-3’; Reverse, 5’-TACCG ATGTC TGGTA GACGAT-3’). PCR conditions were: 55°C 1 min, followed by 35 cycles each containing 92°C denature for 30 sec, 58°C denature for 45 sec and 72°C elongation for 35 sec. Fluorescent quantitative PCR cycler (PE, US) was used to calculate CT values of all samples and standard reference. The expression level of target gene was analyzed by 2-ΔCT method.
MTT assay for cell proliferation
Log-phased PANC-1 cells (5×103 per mL) were inoculated into 96-well plate using DMEM containing 10% fetal bovine serum (FBS). After 24-hour incubation, 20 μL MTT reagents (Sigma, US) were added into each well. After 4-hour incubation, supernatants were removed and 0.15 mL DMSO (Sigma, US) was added. The plate was vibrated for 10 min until complete resolution of violet crystals. The optical absorbance (A) value was quantified form a microplate reader. Growth inhibition rate = (1-mean A value of experimental group/mean A value of control group) ×100%. Triplicated experiments were performed at each time point.
Cell invasion assay by transwell
48 hours after transfection, all cells were cultured in serum-free medium for further 24 hours. Both bottom and upper membrane surface of Transwell chamber (Hyclone, US) were coated with 1:5 Matrigel dilution (50 mg/L) and air-dried. Transwell chambers were put into 24-well plate, with the addition of 0.5 mL DMEM medium containing 0.5 mL 10% FBS. Within each chamber, 0.1 mL tumor cell suspension was seeded with serum-free medium. Meanwhile, equal volumes of cells were added into Transwell chamber with no Matrigel treatment as an internal control. After the 48-hour incubation, Transwell chambers were removed and rinsed in PBS. After the cleaning of cells on the upper surface of the membrane, the whole chamber was fixed in cold ethanol, followed by crystal violet staining for 30 min. The number of cells on the lower surface of the membrane was counted under an inverted microscope. The average number of invasion cells in 10 randomly selected fields was calculated. Triplicated experiments were performed at each time point.
Western blotting
Total proteins were extracted from all cells by lysis buffer incubation on ice (15~30 min), ultrasonic rupture (5 sec ×4) and centrifugation (10000 g, 15 min). The supernatants were transferred to new tubes for quantification. Proteins were firstly separated by 10% SDS-PAGE and transferred to PVDF membrane (Pall Life Sciences, US). Nonspecific binding was blocked by 5% defatted milk powder for 2 hours. Anti-caspase 3 monoclonal antibody (1:500, Cell Signaling, US) was added for overnight incubation, followed by PBST washing and 30-min incubation with mouse anti-rabbit secondary antibody conjugated with horseradish peroxidase (HRP) (1:2000, Cell Signaling, US). The membrane was finally developed using ECL reagent (Amersham Bioscience, US) and exposed under X-ray. Quantity One software was used for calculating optical density of protein bands. A parallel internal control was performed using actin. All experiments were repeated for four times.
Statistical analysis
SPSS 16.0 software package was used to analyze all collected data, of which measurement data were presented as mean ± standard deviation (SD). Between-group-comparison was performed by LSD test. A statistical analysis was defined when P<0.05.
Results
Expression of miR-184 in PNAC-1 and HPDE6c7 cells
Using real-time PCR, we quantified the expressional profile of miR-184 in PDAC cell line PANC-1 and normal pancreatic epithelial cell line HPDE6c7. We found that the expression of miR-184 was significantly increased in PANC-1 cells when compared to HPDE6c7 (P<0.05, Figure 1). The transfection of miR-184 inhibitor into PANC-1 cells could significantly suppress miR-184 expression (P<0.05, Figure 1).
Figure 1.
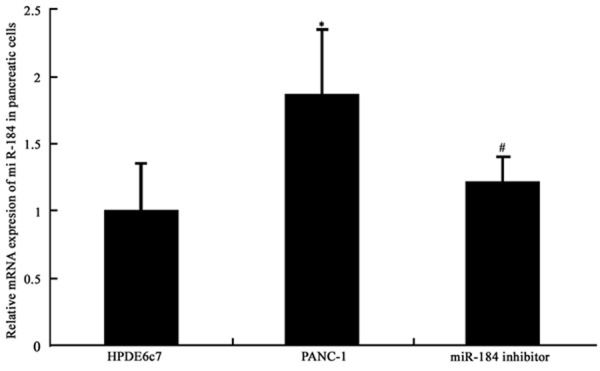
miR-184 expression levels. *P<0.05 compared to HPDE6c7 controlled cells; #P<0.05 compared to PANC-1 cells.
Effect of miR-184 on PANC-1 cell proliferation
MTT assay was used to detect the effect of miR-184 on PANC-1 cell proliferation. We found that after miR-184 inhibition, the proliferation rate of PANC-1 cell was significantly suppressed when compared to control group (P<0.05, Figure 2). This result suggested the facilitating role of miR-184 in the proliferation of PANC-1 cells. The inhibition of miR-184 expression, therefore, can suppress tumor cell proliferation.
Figure 2.
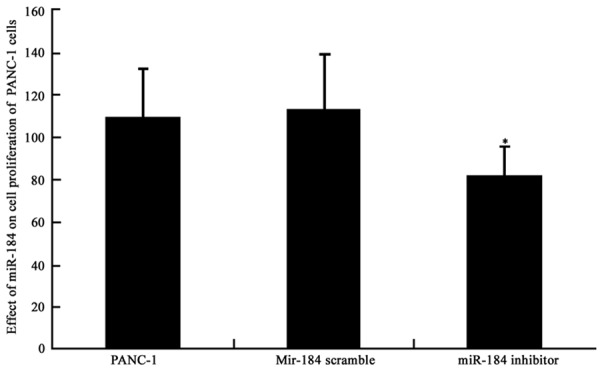
PANC-1 cell proliferation and miR-184 expression. *P<0.05 compared to PANC-1 cells.
Cell invasion ability of PANC-1 cells
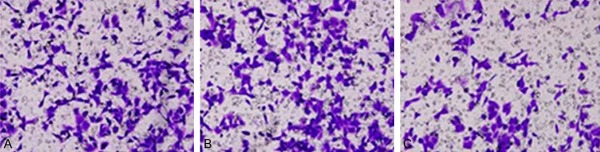
Transwell chamber was employed to test the effect of miR-184 on the invasion ability of PANC-1 cells. The transfection of miR-184 inhibitor significantly decreased invasion ability, as supported by the lower perforated cell number (81±14) when compared to PANC-1 group (121±12), with significant difference (P<0.05, Figures 3, 4). This result suggests the participation of miR-184 in regulating PANC-1 cell proliferation.
Figure 3.
Transwell cell invasion image. A. PANC-1 group; B. Scramble RNA-transfection PANC-1 group; C. miR-184 inhibitor transfection group.
Figure 4.
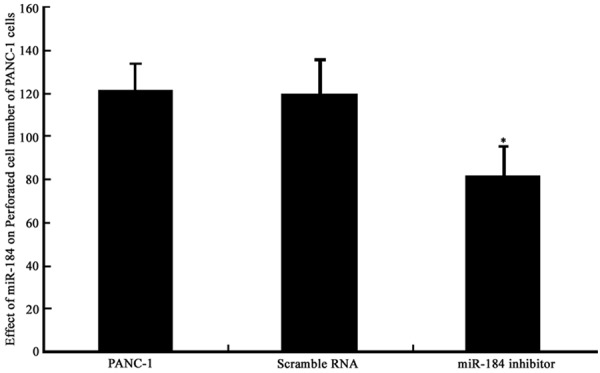
PANC-1 cell invasion ability. *P<0.05 compared to PANC-1 cells.
Apoptotic protein caspase-3 expression
To further illustrate the effect of miR-184 in PANC-1 cells, Western blotting assay was used to describe its expressional profile. Results showed that, after miR-184 transfection, caspase-3 protein was significantly up-regulated in PANC-1 cells when compared to PANC-1 cells (P<0.05, Figures 5, 6). These results indicate the involvement of miR-184 in apoptosis of PANC-1 cells.
Figure 5.
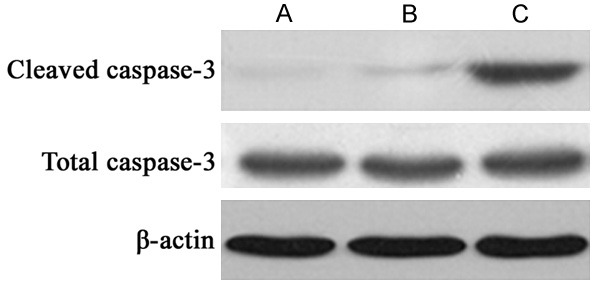
Caspase-3 protein expression. A. PANC-1 group; B. Scramble RNA-transfection PANC-1 group; C. miR-184 inhibitor transfection group.
Figure 6.
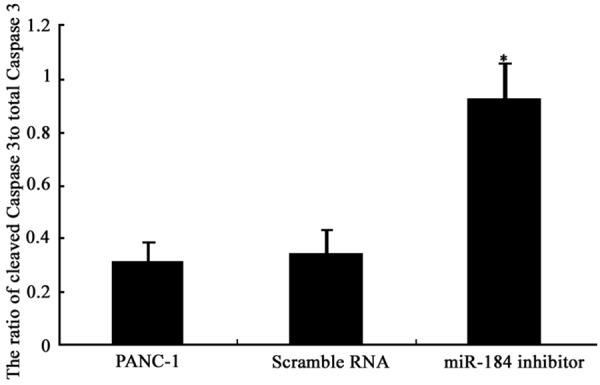
Cleaved caspase-3/total caspase-3 ratio. *P<0.05 compared to PANC-1 cells.
Discussion
Due to its high malignancy and rapid progression, PC is often to be diagnosed until reaching terminal stage, accompanying with metastasis and multi-organ failure. The impossibility for surgical resection makes the 1-year survival rate as high as 65%. Most PC cases are derived from epithelial cells of pancreatic duct, namely PDAC, which has an even more aggressive and unfavorable prognosis compared to other types of PC [15,16]. Currently no reliable biomarker has been identified for early diagnosis of PDAC, further compromising its treatment efficacy [17]. Therefore, the illustration of molecular mechanism underlying the pathogenesis and progression of PDAC can benefit the early diagnosis and treatment of PC, thus improving patients’ life quality and prognosis.
MiRNA, since its first identification in nematode, has been revealed to have a wide array of biological functions including cell proliferation/apoptosis, signaling transduction, tissue differentiation, hormone secretion, lipid metabolism and maintain pluripotency of stem cells. Recent studies have also found the involvement of miRNAs in tumor occurrence, progression, invasion, metastasis and other biological features [18]. Differential expression and biological functions of miRNAs exist in various tumor cells. The expressional profile of miRNA can help to illustrate its relationship with tumor occurrence, progression and differentiation, thus recognizing low-differentiated tumors and benefiting diagnosis and subtyping [19,20]. MiRNA has been found to participate in the regulation of PDAC, as miR-10b can inhibit PDAC cell growth and has been found to be down-regulated in PDAC tissues, suggesting its tumor-suppressor gene-like function, while up-regulation of miR-301a facilitated PDAC cell proliferation [21].
As one newly discovered miRNA family member, miR-184 has been found to be abnormally expressed in various tumor cells, suggesting it potency in regulating oncognenesis and as the target for tumor drugs. The expressional profile of miR-184 in different tumors, however, remained inconsistent. It is up-regulated in glioma tissues, for which it may have potentiated function for tumor occurrence and development, while in breast cancer it works as one tumor-suppressor biomarker [22,23]. Its expression and function in PDAC or normal pancreatic cells remained unclear. This study found up-regulation of miR-184 in PDAC cells compared to normal pancreatic cells. Further transfection using miR-184 inhibitor found inhibited cell proliferation, lower invasion, and up-regulation of apoptotic protein caspase-3 in transfected PANC-1 cells.
In summary, the elevated expression of miR-184 in PDAC can facilitate proliferation and invasion of tumor cells, suppress cell apoptosis, thus promoting the occurrence and progression of PDAC. MiR-184, therefore, has the potency as a novel target for molecular therapy against PDAC, although its detailed mechanism needs further elucidation.
Disclosure of conflict of interest
None.
References
- 1.Singh D, Upadhyay G, Srivastava RK, Shankar S. Recent advances in pancreatic cancer: biology, treatment, and prevention. Biochim Biophys Acta. 2015;1856:13–27. doi: 10.1016/j.bbcan.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Meachem MD, Snead ER, Kidney BA, Jackson ML, Dickinson R, Larson V, Simko E. A comparative proteomic study of plasma in feline pancreatitis and pancreatic carcinoma using 2-dimensional gel electrophoresis to identify diagnostic biomarkers: A pilot study. Can J Vet Res. 2015;79:184–9. [PMC free article] [PubMed] [Google Scholar]
- 3.Takano H, Tsuchikawa T, Nakamura T, Okamura K, Shichinohe T, Hirano S. Potential risk of residual cancer cells in the surgical treatment of initially unresectable pancreatic carcinoma after chemoradiotherapy. World J Surg Oncol. 2015;13:209. doi: 10.1186/s12957-015-0617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taher MA, Khan ZR, Chowdhury MM, Nur-E-Elahi M, Chowdhury AK, Faruque MS, Wahiduzzaman M, Haque MA. Pylorus Preserving Pancreaticoduodenectomy vs. Standard Whipple’s Procedure in Case of Carcinoma head of the Pancreas and Periampullary Carcinoma. Mymensingh Med J. 2015;24:319–25. [PubMed] [Google Scholar]
- 5.Dimastromatteo J, Houghton JL, Lewis JS, Kelly KA. Challenges of Pancreatic Cancer. Cancer J. 2015;21:188–93. doi: 10.1097/PPO.0000000000000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao R, Yang J, Zhou BY, Li DW, Huang P, Luo SQ, Du CY. Conditional survival of pancreatic ductal adenocarcinoma in surgical and nonsurgical patients: a retrospective analysis report from a single institution in China. World J Surg Oncol. 2015;13:196. doi: 10.1186/s12957-015-0608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saadatian Z, Masotti A, Nariman Saleh Fam Z, Alipoor B, Bastami M, Ghaedi H. Single-Nucleotide Polymorphisms Within MicroRNAs Sequences and Their 3’ UTR Target Sites May Regulate Gene Expression in Gastrointestinal Tract Cancers. Iran Red Crescent Med J. 2014;16:e16659. doi: 10.5812/ircmj.16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orang AV, Barzegari A. MicroRNAs in colorectal cancer: from diagnosis to targeted therapy. Asian Pac J Cancer Prev. 2014;15:6989–99. doi: 10.7314/apjcp.2014.15.17.6989. [DOI] [PubMed] [Google Scholar]
- 9.Gandhi NS, Tekade RK, Chougule MB. Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release. 2014;194:238–56. doi: 10.1016/j.jconrel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallach S, Calabuig-Fariñas S, Jantus-Lewintre E, Camps C. MicroRNAs: promising new antiangiogenic targets in cancer. Biomed Res Int. 2014;2014:878450. doi: 10.1155/2014/878450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindner K, Haier J, Wang Z, Watson DI, Hussey DJ, Hummel R. Circulating microRNAs: emerging biomarkers for diagnosis and prognosis in patients with gastrointestinal cancers. Clin Sci (Lond) 2015;128:1–15. doi: 10.1042/CS20140089. [DOI] [PubMed] [Google Scholar]
- 12.Cui QK, Liu WD, Zhu JX, Wang YH, Wang ZG. MicroRNA-184 promotes proliferation ability of glioma cells by regulating FOXO3. Asian Pac J Trop Med. 2014;7:776–9. doi: 10.1016/S1995-7645(14)60135-8. [DOI] [PubMed] [Google Scholar]
- 13.Wong TS, Ho WK, Chan JY, Ng RW, Wei WI. Mature miR-184 and squamous cell carcinoma of the tongue. Scientific World Journal. 2009;9:130–2. doi: 10.1100/tsw.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JX, Gao J, Ding SL, Wang K, Jiao JQ, Wang Y, Sun T, Zhou LY, Long B, Zhang XJ, Li Q, Liu JP, Feng C, Liu J, Gong Y, Zhou Z, Li PF. Oxidative Modification of miR-184 Enables It to Target Bcl-xL and Bcl-w. Mol Cell. 2015;59:50–61. doi: 10.1016/j.molcel.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Corapcioglu ME, Ogul H. miSEA: microRNA set enrichment analysis. Biosystems. 2015;134:37–42. doi: 10.1016/j.biosystems.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Bledsoe JR, Shinagare SA, Deshpande V. Difficult Diagnostic Problems in Pancreatobiliary Neoplasia. Arch Pathol Lab Med. 2015;139:848–57. doi: 10.5858/arpa.2014-0205-RA. [DOI] [PubMed] [Google Scholar]
- 17.Galvan JA, Zlobec I, Wartenberg M, Lugli A, Gloor B, Perren A, Karamitopoulou E. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br J Cancer. 2015;112:1944–50. doi: 10.1038/bjc.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varshney J, Subramanian S. MicroRNAs as potential target in human bone and soft tissue sarcoma therapeutics. Front Mol Biosci. 2015;2:31. doi: 10.3389/fmolb.2015.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Yang H. Integrated analysis of differentially expressed genes in breast cancer pathogenesis. Oncol Lett. 2015;9:2560–2566. doi: 10.3892/ol.2015.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preis M, Gardner TB, Gordon SR, Pipas JM, Mackenzie TA, Klein EE, Longnecker DS, Gutmann EJ, Sempere LF, Korc M. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2011;17:5812–21. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bera A, VenkataSubbaRao K, Manoharan MS, Hill P, Freeman JW. A miRNA signature of chemoresistant mesenchymal phenotype identifies novel molecular targets associated with advanced pancreatic cancer. PLoS One. 2014;9:e106343. doi: 10.1371/journal.pone.0106343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phua YW, Nguyen A, Roden DL, Elsworth B, Deng N, Nikolic I, Yang J, Mcfarland A, Russell R, Kaplan W, Cowley MJ, Nair R, Zotenko E, O’Toole S, Tan SX, James DE, Clark SJ, Kouros-Mehr H, Swarbrick A. MicroRNA profiling of the pubertal mouse mammary gland identifies miR-184 as a candidate breast tumour suppressor gene. Breast Cancer Res. 2015;17:83. doi: 10.1186/s13058-015-0593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao B, Gao K, Li L, Huang Z, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC) Biomed Pharmacother. 2014;68:143–8. doi: 10.1016/j.biopha.2013.09.005. [DOI] [PubMed] [Google Scholar]