Abstract
Objectives: Our study was aimed to make sure whether ADAM12 could serve as a prognostic biomarker of estrogen receptor (ER) -positive breast cancer. Methods: 127 patients with ER-positive breast cancer were included in the present study. The level of ADAM12 was assayed through real-time quantitative PCR (RT-qPCR). Levels of ADAM12 in tumor tissues and adjacent normal tissues were compared with paired t-test. The association of ADAM12 expression with clinical characteristics was analyzed via χ2 test. Kaplan-Meier survival curve was used to evaluate the role of ADAM12 expression in overall survival (OS) of patients. Cox-regression analysis was performed to judge if ADAM12 could serve as a prognostic marker in breast cancer. Results: The level of ADAM12 was upregulated in tumor tissues of breast cancer compared to that of adjacent normal tissues (P < 0.05). The expression of ADAM12 was closely related to the Ki-67 and HER2 status (P < 0.05 for both). The results of Kaplan-Meier survival curve showed that patients with higher level of ADAM12 exhibited shorter survival time compared to that of low level of ADAM12 (P < 0.001). Cox regression analysis showed that ADAM12 might be a biomarker in predicting prognosis of patients with ER-positive breast cancer (HR = 7.116, 95% CI = 3.329-15.212). Conclusion: ADAM12 appears to be a prognostic marker in ER-positive breast cancer.
Keywords: ADAM12, clinical outcome, estrogen receptor, breast cancer
Introduction
Breast cancer is a leading cause of cancer-related death in women [1], which is characterized by diverse clinical behavior and outcomes [2]. The newly diagnosed patients are commonly presented with early-stage breast cancer, however 20% of them will suffer recurrence in 10 years after diagnosis [3]. In recent years, despite the advances in early diagnosis and treatment therapy, there are still many patients died from cancer recurrence and metastasis. Until now, there has been many studies that proposed biomarkers for tumor behavior and clinical outcome in breast cancer [4-8], however, the identification of new markers is still necessary [9]. Among the factors related with cancer progression, abnormal genes expression has attracted a lot of attention, some of which have been demonstrated as independent biomarkers for breast cancer.
ADAMs, a group of membrane-associated metalloproteinases, plays an important role in regulating integrin-mediated cell adhesion, cell signal transduction and proteolytic release [10-12]. The family contains 20 members, 9 out of which are produced beyond the reproductive system: ADAM8, -9, -10, -12, -15, -17, -19, -28 and -33. The expression of ADAM12 is upregulated in various cancers [13-15]. Moreover, ADAM12 has been demonstrated as a susceptibility gene of breast cancer [16]. Fröhlich et al. reported that overexpression of ADAM12 accelerated the progression of breast cancer [17]. The related study also suggested that the level of ADAM12 was associated with disease status and pathologic stage in breast cancer [14,17]. However, there are no studies investigating the effects of ADAM12 expression on the survival of patients with breast cancer.
Our study was aimed to ensure if ADAM12 expression served as a prognostic biomarker in breast cancer. The expression level of ADAM12 in serum and tissues (tumor tissues and adjacent normal tissues) were tested with real-time quantitative PCR (RT-qPCR). Kaplan-Meier and Cox analyses were conducted to the role of ADAM12 in breast cancer.
Materials and methods
Participants
In this research, 205 women from Affiliated Central Hospital of Huzhou Teachers College with breast cancer were selected. Among them, 76 individuals were diagnosed as estrogen receptor (ER) negative and 129 were ER positive. 129 women with ER positive breast cancer were included. They were aged 25-75 years old at the time of diagnosis. The patients who had metastatic disease, tumor history and received neoadjuvant chemotherapy were excluded. After the selection, 127 patients were selected.
Pathological information was collected: primary tumor size, Ki-67 status and HER2 status. Before surgery, the blood samples of each individual were collected. Meanwhile, 127 tumor samples and adjacent normal samples were also collected for further analysis. The follow-ups were performed after the treatments. In the survey, survival time, date of death and date of last follow-up were recorded. The overall survival time was defined from the time as treatment began to death time or date of last follow-up.
All the participants signed the written consent before the study and the study was approved by The Ethical Committee of the hospital.
RT-qPCR assay
Total RNA was extracted from serum and tissue samples by Qiagen DNeasy Blood & Tissue Kit, followed by cDNA synthesis with First Strand cDNA Synthesis Kit (Thermo Scientific). GAPDH was used as internal control. The expression of ADAM12 was analyzed with Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The adopted calculation method was 2-ΔΔCT. All the samples were tested in triplicate.
Statistics
All the analysis was completed in SPSS 12.0. The differences in the level of ADAM12 between tumor tissues and adjacent normal tissues were compared with paired t-test. The relationship of expression of ADAM12 with clinical features was analyzed via χ2 test. Kaplan-Meier analysis was adopted to evaluate the effects of ADAM12 expression on the overall survival (OS) of patients with breast cancer. Whether ADAM12 could serve as a prognostic marker in breast cancer was estimated by Cox-regression analysis. The figures were finished in Graphpad prism 5. All the tests were two-tailed, and P < 0.05 indicated that there was significant difference.
Results
The expression level of ADAM12 in serum and tissues
The level of ADAM12 in serum and tissues were tested with RT-qPCR technology. The relative content of ADAM12 in serum was 3.74. It was obvious that ADAM12 expression level was higher in tumor tissues compared to that of adjacent normal tissues (P < 0.05) (Figure 1).
Figure 1.
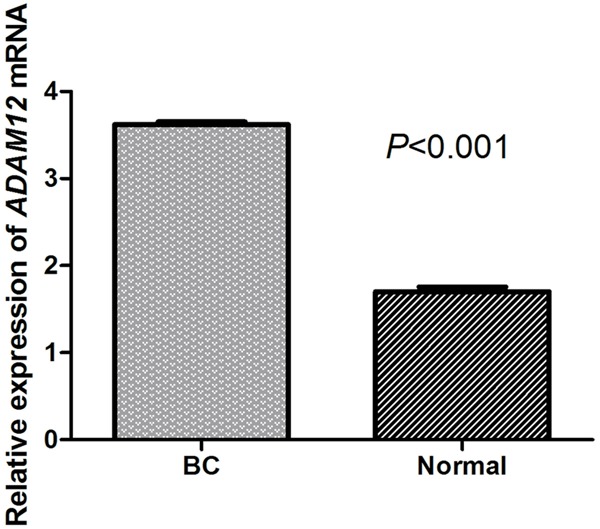
The expression level of ADAM12 in tumor tissues and adjacent normal tissues of ER-positive breast cancer. The level of ADAM12 in tumor tissues was significantly higher than that of normal tissues (P < 0.001). BC indicates breast cancer.
Basic characteristic of patients with ER-positive breast cancer
The clinical characteristics of patients with ER-positive breast cancer were listed in Table 1. The patients were divided into two groups according to the level of ADAM12. There were no obvious differences in age, family history and tumor size between two groups (P > 0.05 for all). While, it turned out that ADAM12 level was significantly related with Ki-67 and HER2 status (P < 0.05 for both), which indicates that ADAM12 may be correlated with prognosis in breast cancer.
Table 1.
Relationship between ADAM12 expression and clinical features of patients
| Characteristics | Case NO. | Expression | χ2 | P value | |
|---|---|---|---|---|---|
|
| |||||
| High | Low | ||||
| Age | 0.961 | 0.327 | |||
| ≤ 49 | 60 | 41 | 19 | ||
| > 49 | 67 | 51 | 16 | ||
| Family history | 1.298 | 0.255 | |||
| Yes | 72 | 55 | 17 | ||
| No | 55 | 37 | 18 | ||
| Tumor size | 0.339 | 0.561 | |||
| ≤ 3.5 | 82 | 58 | 24 | ||
| > 3.5 | 45 | 34 | 11 | ||
| Ki-67 | 4.958 | 0.026 | |||
| Positive | 71 | 57 | 14 | ||
| Negative | 56 | 35 | 21 | ||
| HER2 | 4.013 | 0.045 | |||
| High | 76 | 60 | 16 | ||
| Low | 51 | 32 | 19 | ||
Kaplan-Meier survival curve and Cox regression analysis
Kaplan-Meier survival curve was constructed to determine the differences in survival situation of two patients. The result illustrated that high level of ADAM12 predicted worse prognosis of patients with ER-positive breast cancer (P < 0.001) (Figure 2). Further analysis of Cox regression provided evidence that ADAM12 was a promising prognostic biomarker in ER-positive breast cancer (HR = 7.116, 95% CI = 3.329-15.212) (Table 2).
Figure 2.
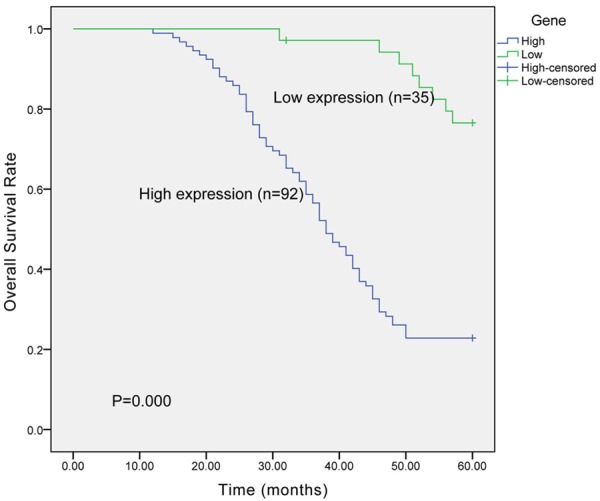
Kaplan-Meier survival curve. Increased expression of ADAM12 predicted worse clinical outcome of patients with ER-positive breast cancer.
Table 2.
Multivariate analysis of ADAM12 expression in ER-positive breast cancer
| Characteristics | P value | HR | 95% CI |
|---|---|---|---|
| Age | 0.161 | 0.725 | 0.462-1.137 |
| Tumor size | 0.145 | 0.691 | 0.420-1.136 |
| Ki-67 | 0.347 | 1.275 | 0.768-2.116 |
| HER2 | 0.615 | 1.132 | 0.698-1.835 |
| ADAM12 expression | 0.000 | 7.116 | 3.329-15.212 |
Discussion
ADAMs have specific extracellular domains, such as a prodomain, metalloproteinases domain, epidermal growth factor (EGF)-like repeat domain, disintegrin domain, cysteine-rich domain, transmembrane domain and cytoplasmic domain. ADAMs show many important biological activities through these domains. First, ADAMs can shed growth factors of TGF-α and HB-EGF, which results in promoted cell proliferation. Second, ADAMs serve as adhesion molecules by binding to inregrins with cysteine-rich and disintegrin domains. Third, ADAMs incline to regulate cell proliferation signals via integrins. Fourth, the proteinase activity of ADAMs to membrane-anchored molecules of cytokines, chemokines or its receptors laid basis for its role in cancer progression. The fact is that ADAMs contribute to the occurrence of many cancers [18].
ADAM12 is a member of ADAM family. It has certain crucial biological functions in tumors. Like other ADAMs, ADAM12 also could regulate proteolytic ectodomain and result in shedding of growth factors [11,19,20]. Besides, it is involved in nonproteolytic protein-protein interactions. The expression level of ADAM12 is always low. Nevertheless, its increased level was observed in liver cancer, bladder cancer, lung cancer and breast cancer [21-24]. ADAM12 is commonly expressed as two forms: transmembrane form (ADAM12-L) and secreted form (ADAM12-S). The research of Roy et al. found that overexpression of ADAM12-L and ADAM12-S in breast cancer cells could promote the estrogen-independent proliferation [25].
In terms of the abnormality behavior of ADAM12 in cancers or diseases, there has been many studies investigating the specific role in single cancer or disease. Yang et al. concluded that ADAM12 was a promising diagnostic marker for complete spontaneous abortion and ectopic pregnancy in symptomatic women [26]. A study on oral squamous cell carcinoma indicated that high level of ADAM12 could accelerate cell proliferation [27]. The positive activation loop between ADAM12 and HER2 may contribute to tumor cell migration and invasion in human head and neck cancer [28]. It also appears to be a prognostic marker in resected pethological stage I lung adenocarcinoma [23].
Based on the previous studies, our study tested the expression level of ADAM12 in serum and tissues. The results showed that the level of ADAM12 was upregulated in tumors serum and tissues. Moreover, its expression was associated with Ki-67 and HER2 status. The Kaplan-Meier survival curve illustrated that the patients with high level ADAM12 were more likely to experience short survival time. Cox regression analysis indicated that ADAM12 was an independent marker for predicting clinical outcome in breast cancer.
Taken together, ADAM12 may serve as a promising biomarker for predicting prognosis of patients with breast cancer. The present study only considered the effects of single gene on the survival situation, future researches should take more factors into account to get better understand on the pathological mechanism of breast cancer. ADAMs play its roles by many ways, so further studies to make sure the function mechanism of ADAM12 in breast cancer are necessary for promoting the treatments of patients.
Disclosure of conflict of interest
None.
References
- 1.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2014. Ann Oncol. 2014;25:1650–1656. doi: 10.1093/annonc/mdu138. [DOI] [PubMed] [Google Scholar]
- 2.Cadoo KA, Fornier MN, Morris PG. Biological subtypes of breast cancer: current concepts and implications for recurrence patterns. Q J Nucl Med Mol Imaging. 2013;57:312–321. [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M, Larsen MJ, Kruse TA. Long non-coding RNA expression profiles predict metastasis in lymph node-negative breast cancer independently of traditional prognostic markers. Breast Cancer Res. 2015;17:55. doi: 10.1186/s13058-015-0557-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka M, Ichikawa-Tomikawa N, Shishito N, Nishiura K, Miura T, Hozumi A, Chiba H, Yoshida S, Ohtake T, Sugino T. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer. 2015;15:53. doi: 10.1186/s12885-015-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akter S, Choi TG, Nguyen MN, Matondo A, Kim JH, Jo YH, Jo A, Shahid M, Jun DY, Yoo JY, Ngo YN, Seo SW, Ali L, Lee JS, Yoon KS, Choe W, Kang I, Ha J, Kim J, Kim SS. Prognostic value of a 92-probe signature in breast cancer. Oncotarget. 2015;6:15662–80. doi: 10.18632/oncotarget.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedau S, Batra M, Singh UR, Bharti AC, Ray A, Das BC. Expression of BRCA1 and BRCA2 proteins and their correlation with clinical staging in breast cancer. J Cancer Res Ther. 2015;11:158–163. doi: 10.4103/0973-1482.140985. [DOI] [PubMed] [Google Scholar]
- 8.Cheng CL, Thike AA, Tan SY, Chua PJ, Bay BH, Tan PH. Expression of FGFR1 is an independent prognostic factor in triple-negative breast cancer. Breast Cancer Res Treat. 2015;151:99–111. doi: 10.1007/s10549-015-3371-x. [DOI] [PubMed] [Google Scholar]
- 9.Banin Hirata BK, Oda JM, Losi Guembarovski R, Ariza CB, de Oliveira CE, Watanabe MA. Molecular markers for breast cancer: prediction on tumor behavior. Dis Markers. 2014;2014:513158. doi: 10.1155/2014/513158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blobel CP, Carpenter G, Freeman M. The role of protease activity in ErbB biology. Exp Cell Res. 2009;315:671–682. doi: 10.1016/j.yexcr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 12.Murphy G. Fell-Muir Lecture: Metalloproteinases: from demolition squad to master regulators. Int J Exp Pathol. 2010;91:303–313. doi: 10.1111/j.1365-2613.2010.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iba K, Albrechtsen R, Gilpin BJ, Loechel F, Wewer UM. Cysteine-rich domain of human ADAM 12 (meltrin alpha) supports tumor cell adhesion. Am J Pathol. 1999;154:1489–1501. doi: 10.1016/s0002-9440(10)65403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kveiborg M, Frohlich C, Albrechtsen R, Tischler V, Dietrich N, Holck P, Kronqvist P, Rank F, Mercurio AM, Wewer UM. A role for ADAM12 in breast tumor progression and stromal cell apoptosis. Cancer Res. 2005;65:4754–4761. doi: 10.1158/0008-5472.CAN-05-0262. [DOI] [PubMed] [Google Scholar]
- 15.Tian B, Wen J, Zhang M, Li Q, Xie D. [Expression of ADAM12 (Meltrin-alpha) gene in giant cell tumor of bone] . Zhonghua Bing Li Xue Za Zhi. 2001;30:350–352. [PubMed] [Google Scholar]
- 16.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 17.Frohlich C, Nehammer C, Albrechtsen R, Kronqvist P, Kveiborg M, Sehara-Fujisawa A, Mercurio AM, Wewer UM. ADAM12 produced by tumor cells rather than stromal cells accelerates breast tumor progression. Mol Cancer Res. 2011;9:1449–1461. doi: 10.1158/1541-7786.MCR-11-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy MJ, Mullooly M, O’Donovan N, Sukor S, Crown J, Pierce A, McGowan PM. The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer? Clin Proteomics. 2011;8:9. doi: 10.1186/1559-0275-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40:1685–1702. doi: 10.1016/j.biocel.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Jacobsen J, Wewer UM. Targeting ADAM12 in human disease: head, body or tail? Curr Pharm Des. 2009;15:2300–2310. doi: 10.2174/138161209788682389. [DOI] [PubMed] [Google Scholar]
- 21.Le Pabic H, Bonnier D, Wewer UM, Coutand A, Musso O, Baffet G, Clement B, Theret N. ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37:1056–1066. doi: 10.1053/jhep.2003.50205. [DOI] [PubMed] [Google Scholar]
- 22.Frohlich C, Albrechtsen R, Dyrskjot L, Rudkjaer L, Orntoft TF, Wewer UM. Molecular profiling of ADAM12 in human bladder cancer. Clin Cancer Res. 2006;12:7359–7368. doi: 10.1158/1078-0432.CCR-06-1066. [DOI] [PubMed] [Google Scholar]
- 23.Mino N, Miyahara R, Nakayama E, Takahashi T, Takahashi A, Iwakiri S, Sonobe M, Okubo K, Hirata T, Sehara A, Date H. A disintegrin and metalloprotease 12 (ADAM12) is a prognostic factor in resected pathological stage I lung adenocarcinoma. J Surg Oncol. 2009;100:267–272. doi: 10.1002/jso.21313. [DOI] [PubMed] [Google Scholar]
- 24.Lendeckel U, Kohl J, Arndt M, Carl-McGrath S, Donat H, Rocken C. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J Cancer Res Clin Oncol. 2005;131:41–48. doi: 10.1007/s00432-004-0619-y. [DOI] [PubMed] [Google Scholar]
- 25.Roy R, Moses MA. ADAM12 induces estrogen-independence in breast cancer cells. Breast Cancer Res Treat. 2012;131:731–741. doi: 10.1007/s10549-011-1431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Wu J, Guo F, Wang D, Chen K, Li J, Du L, Yin A. Maternal serum disintegrin and metalloprotease protein-12 in early pregnancy as a potential marker of adverse pregnancy outcomes. PLoS One. 2014;9:e97284. doi: 10.1371/journal.pone.0097284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uehara E, Shiiba M, Shinozuka K, Saito K, Kouzu Y, Koike H, Kasamatsu A, Sakamoto Y, Ogawara K, Uzawa K, Tanzawa H. Upregulated expression of ADAM12 is associated with progression of oral squamous cell carcinoma. Int J Oncol. 2012;40:1414–1422. doi: 10.3892/ijo.2012.1339. [DOI] [PubMed] [Google Scholar]
- 28.Rao VH, Kandel A, Lynch D, Pena Z, Marwaha N, Deng C, Watson P, Hansen LA. A positive feedback loop between HER2 and ADAM12 in human head and neck cancer cells increases migration and invasion. Oncogene. 2012;31:2888–2898. doi: 10.1038/onc.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]


