Figure 4.
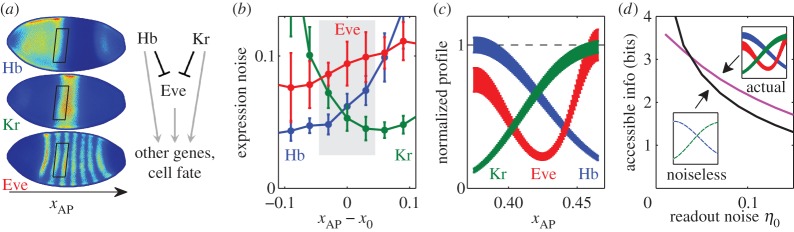
(a) Immunostaining of three AP axis patterning genes in the same embryo. Rather than specifying cell fate directly, the ‘gap genes’ such as hunchback (Hb; top) and Krüppel (Kr; middle) control ‘pair-rule’ genes such as even-skipped (Eve, bottom). Both tiers regulate other genes further downstream. Boxes indicate the selected ROI, where at this time, Hb and Kr are the only relevant inputs to Eve, as shown in the cartoon. (b) Within the ROI (shaded), Eve exhibits higher expression noise than either Hb or Kr. Expression noise computed as RMS difference between expression level of a nucleus and its immediate dorsal or ventral neighbour (see the electronic supplementary material), plotted against AP distance from the Hb/Kr boundary (denoted x0). Error bars are standard deviation over N=8 embryos. (c) Idealized morphogen profiles, restricted to the ROI. Profile shape obtained as smooth spline-fit to expression values and noise magnitudes calculated for the profiles of panel (a) after projection onto the AP axis. (d) For all but the lowest readout noise magnitude, joint accessible information content in the triplet (Hb, Kr, Eve) exceeds the accessible information provided by Hb and Kr alone, even in an extreme hypothetical case when they are rendered entirely noiseless.