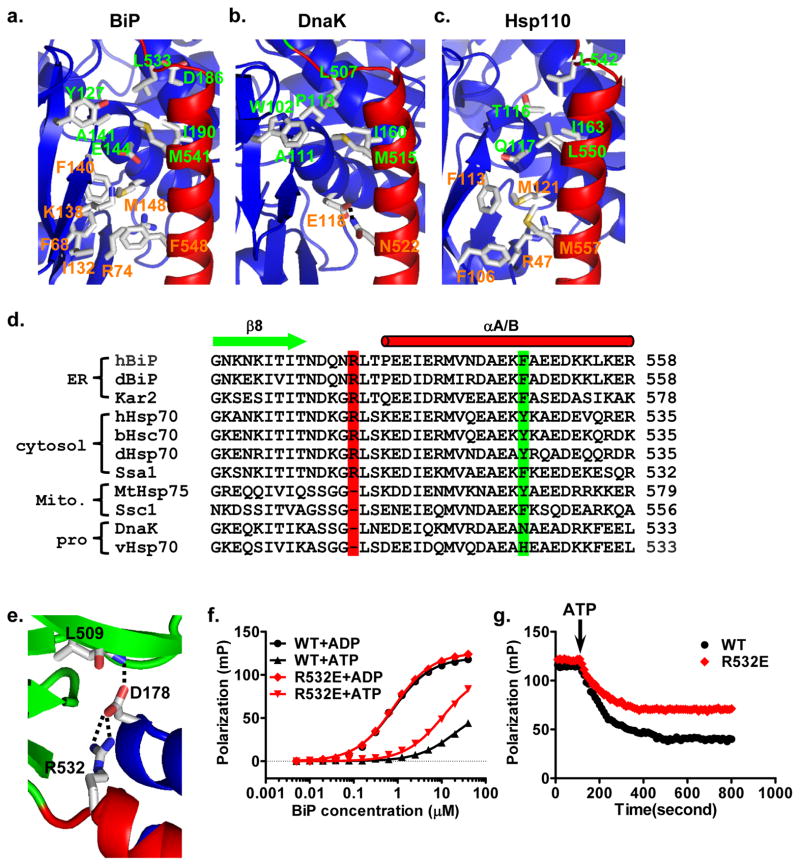
Figure 4. The unique NBD-SBDα interface and NBD-Lα,β contact in BiP-ATP.
a–c, Ribbon diagrams of NBD-SBDα interfaces in the BiP-ATP (a), DnaK-ATP (PDB code: 4JNE) (b), and Sse1-ATP (PDB code: 2QXL) (c) structures. NBDs are in blue and SBDαs are in red. Residues forming the two clusters of contacts are shown in stick presentation. Residues labeled in green are highly conserved between BiP-ATP and DnaK-ATP; residues labeled in orange are conserved between BiP-ATP and Sse1-ATP.
d, Sequence alignment among Hsp70s. Secondary structure assignments are labeled on the top with cylinder for helix and arrow for strand. R532 and F548 are highlighted in red and green, respectively. h, human; d, Drosophila melanogaster; b, bovine; v, Virgibacillus halodenitrificans. DnaK is from E.coli. Kar2, Ssa1 and Ssc1 are from saccharomyces cerevisiae.
e, The unique contact of NBD-Lα,β in the BiP-ATP structure. R532 forms two hydrogen bonds with D178 on NBD (blue). SBDβ and SBDα are in green and red, respectively.
f, Fluorescence anisotropy assay of NR peptide binding affinity for BiP R532E mutant. WT BiP was used as a control. Assays were carried out as in Fig. 1 in the presence of ATP (+ATP) or ADP (+ADP).
g, BiP R532E has a defect in releasing NR peptide upon addition of ATP. BiP proteins were incubated with F-NR peptide in the presence of ADP. After binding reached equilibrium, ATP was added (indicated by an arrow), and the release of F-NR was monitored over time.