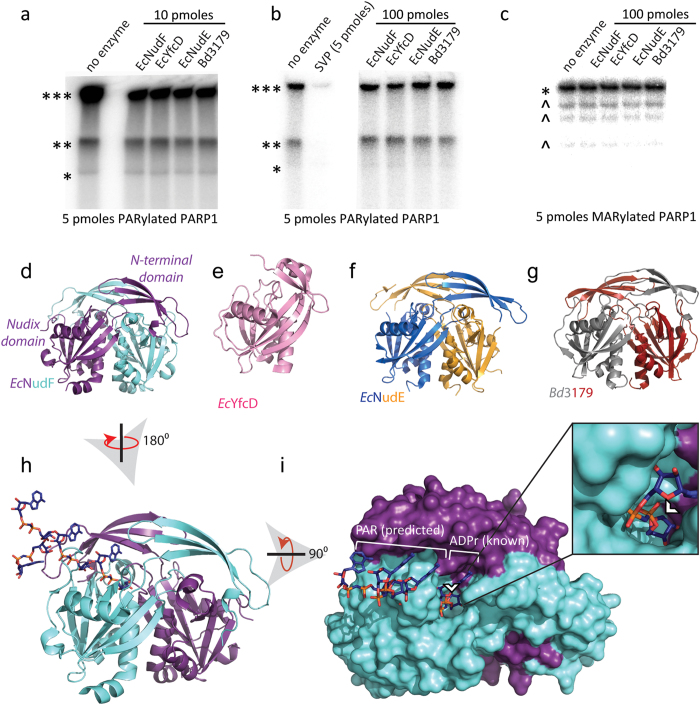
Figure 2. Nudix ADPrases are ineffective against protein-conjugated ADPr.
(a–c) Autoradiograph showing 32P-labeled mono- or poly(ADP-ribose) conjugated to PARP1 following exposure to canonical nucleotide sugar hydrolases: EcNudF, EcYfcD and EcNudE and Bd3179. For (a), 5 pmoles of PARylated PARP1 was exposed to 10 pmoles of hydrolase for 2 hrs at 37 °C; for (b), 100 pmoles of hydrolases were utilized, while in (c) 5 pmoles of PARP1 E988Q, a mutant only capable of synthesizing mono(ADP-ribose), was exposed to 100 pmoles of hydrolases for 2 hrs at 37 °C. Ribbon diagrams show the structure of each of the enzymes used: (d) EcNudF/ADPrase (PDB ID 1KHZ)18, (e) EcYfcD (PDB ID 2FKB), (f) EcNudE (PDB ID 1VHG) and (g) Bd3179 (PDB ID 5C7Q)52. Panel (h) shows a surface representation of EcNudF with modeled PAR polymer based on the complex with a nonhydrolyzable ADPr48. Panel (i) shows a surface representation of EcNudF with the modeled PAR polymer, the inset shows how the terminal ribose is protected and buried within the enzyme. The white arrowhead indicates where protein conjugation would occur on ADPr. ***SDS-PAGE well, **SDS-PAGE interface between stacking and resolving gels, *native size of PARP1, ^co-purified PARP1 protein fragments.