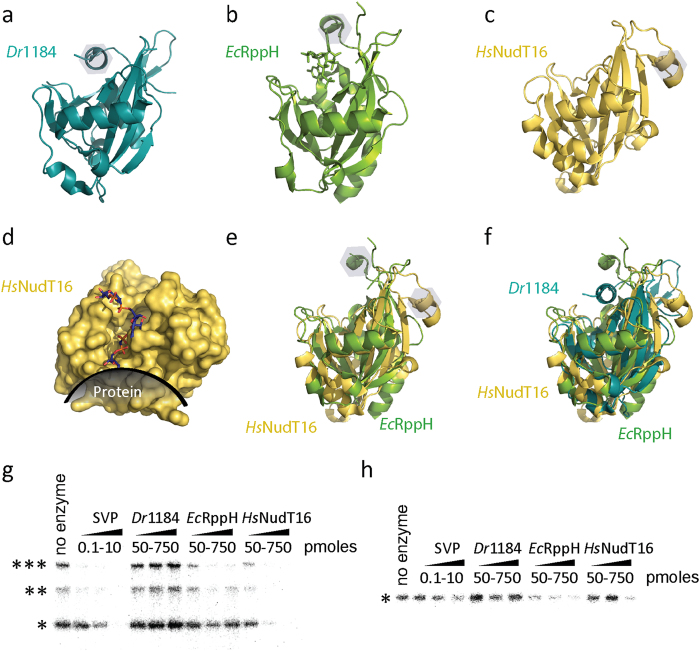
Figure 5. HsNudT16 degrades protein-conjugated ADPr.
(a) A ribbon model of the structure of Dr1184/CoAse (teal, PDB ID 1NQY). (b) A ribbon model of the structure of EcRppH (green, PDB ID 4S2Y). (c) A ribbon model of HsNudT16 (PDB ID 2XSQ), (d) Surface representation of HsNudT16 with PAR modeled (sticks) and a protein depicted at the conjugation site of ADPr. Panel (e) shows a structural alignment between HsNudT16 and EcRppH, while (f) shows a structural alignment of HsNudT16, EcRppH and Dr1184. The grey hexagons in panels (a), (b), (c) and (e) show the helix that moves upon substrate binding. Panels (g) and (h) show the removal of 32P-labeled ADPr from 5 pmoles of PARylated (g) or MARylated (h) PARP1 by increasing amounts of SVP (0.1, 1 and 10 pmoles) or Dr1184, EcRppH or HsNudT16 (50, 250 and 750 pmoles). ***SDS-PAGE well, **SDS-PAGE interface between stacking and resolving gels, *native size of PARP1.