Abstract
Escherichia coli physiological, biomass elemental composition and proteome acclimations to ammonium-limited chemostat growth were measured at four levels of nutrient scarcity controlled via chemostat dilution rate. These data were compared with published iron- and glucose-limited growth data collected from the same strain and at the same dilution rates to quantify general and nutrient-specific responses. Severe nutrient scarcity resulted in an overflow metabolism with differing organic byproduct profiles based on limiting nutrient and dilution rate. Ammonium-limited cultures secreted up to 35 % of the metabolized glucose carbon as organic byproducts with acetate representing the largest fraction; in comparison, iron-limited cultures secreted up to 70 % of the metabolized glucose carbon as lactate, and glucose-limited cultures secreted up to 4 % of the metabolized glucose carbon as formate. Biomass elemental composition differed with nutrient limitation; biomass from ammonium-limited cultures had a lower nitrogen content than biomass from either iron- or glucose-limited cultures. Proteomic analysis of central metabolism enzymes revealed that ammonium- and iron-limited cultures had a lower abundance of key tricarboxylic acid (TCA) cycle enzymes and higher abundance of key glycolysis enzymes compared with glucose-limited cultures. The overall results are largely consistent with cellular economics concepts, including metabolic tradeoff theory where the limiting nutrient is invested into essential pathways such as glycolysis instead of higher ATP-yielding, but non-essential, pathways such as the TCA cycle. The data provide a detailed insight into ecologically competitive metabolic strategies selected by evolution, templates for controlling metabolism for bioprocesses and a comprehensive dataset for validating in silico representations of metabolism.
Introduction
Nutrient scarcity is a common limiter of microbial growth and is therefore a major source of selective pressure. Evolution has selected fit organisms with competitive functionalities including the ability to sense environmental nutrient levels, acquire resources effectively using high-affinity transporters and invest limiting resources into phenotypes that favour growth or persistence. Studying microbial acclimation to nutrient limitation is of general and specific importance to applied biological efforts, including the control of medical pathogens and synthesis of industrial biochemicals.
Chemostats are powerful tools for measuring the interplay between environment and phenotype, and for controlling bioprocesses (Bull, 2010; Ihssen & Egli, 2004; Pirt, 1975). Chemostats represent spatially and temporally uniform ecosystems that can test microbial acclimations to single stressors (Monod, 1950; Novick & Szilard, 1950). Culture-specific growth rate and the magnitude of nutrient-limitation stress are controlled by the chemostat dilution rate. Chemostats avoid some complexities associated with batch culturing, including time-dependent changes in specific growth rates, extracellular substrate availabilities and environmental stresses such as pH; these complications can confound efforts to assign cause and effect to cellular behaviours.
Escherichia coli is currently the most thoroughly studied micro-organism and is a common bioprocess host, yet substantial gaps in fundamental physiological knowledge exist. Glucose-limited growth has been studied extensively (e.g. Hua et al., 2004; Ihssen & Egli, 2004; Neijssel et al., 1996; Senior, 1975), but E. coli acclimation to other common nutrient limitations such as nitrogen or iron is not as well established. Nitrogen is a common limiter of microbial growth in many natural ecosystems and is commonly used in bioprocesses to induce accumulation of biofuels or bioplastics (Keshavarz & Roy, 2010). A number of chemostat studies have reported the effects of nitrogen limitation on E. coli growth (Senior, 1975), fluxome (Hua et al., 2003; Sauer et al., 1999), transcriptome (Sukmarini & Shimizu, 2010) and metabolome (Brauer et al., 2006), although E. coli chemostat studies of nitrogen-limited growth with proteomics analysis are limited in both occurrence and protein coverage. Only four protein expression studies for nitrogen-limited E. coli chemostats were identified: two focused solely on outer membrane proteins (Liu & Ferenci, 1998; Schliep et al., 2012), a third examined nitrogen-assimilating enzyme activity at different dilution rates (Senior, 1975) and the fourth studied the effect of ammonium limitation on only four cytoplasmic proteins (Ihssen & Egli, 2004). The study by Ihssen & Egli (2004) also discovered and discussed probable complications with some earlier chemostat studies; they hypothesized some of these studies with high cell densities were limited for trace metals in addition to the reported limiting nutrient, confounding interpretation of the data. Iron limitation is another important bacterial nutrient-limitation stress. It is central in medical infections and to marine microbial growth, and has been described recently by our group (Folsom et al., 2014).
Biomass elemental composition quantifies the relative cellular requirements for anabolic resources and reflects a broad, systems-wide averaging of macromolecular composition (Sterner & Elser, 2002). Biomass stoichiometry is also representative of gross cellular energetic content as quantified by the degree of reduction (Roels, 1980). The elemental composition of some bacteria has been reported to change as a function of limiting nutrient (Egli, 1991), growth rate (Chrzanowski & Grover, 2008) and growth temperature (Chrzanowski & Grover, 2008; Cotner et al., 2006). Whilst a popular analysis in previous decades, reporting experimental biomass stoichiometry is no longer commonplace outside of ecological studies (e.g. Ho & Payne, 1979; Scott et al., 2012; Simonds et al., 2010). E. coli biomass elemental composition has been reported for some culturing conditions, (Bauer & Ziv, 1976; Cordier et al., 1987; Heldal et al., 1996; Pickett et al., 1979), but none of these studies reported the biomass stoichiometry for ammonium- or iron-limited E. coli growth.
Here, we report E. coli physiology, biomass elemental content and central metabolism proteome for ammonium-limited chemostat growth at four dilution rates, and contrast the data with published iron- and glucose-limited growth at the same dilution rates using the same E. coli K-12 strain, M9 medium and reactor configuration (Folsom et al., 2014). The previously reported data were leveraged to gain additional physiological knowledge by noting nutrient-specific and general stress responses. The nutrients have varied roles in biomass synthesis and energy metabolism. Ammonium is a macronutrient used to synthesize major biomass constituents, including protein, RNA and DNA; iron is an essential micronutrient used primarily as an enzyme cofactor; glucose is both the sole reduced carbon source and the sole energy source, and therefore has critical anabolic and catabolic roles, respectively. The research quantifies microbial acclimation to common nutrient-scarcity stresses, which in turn provides ecological insight into evolutionary selection, provides a dataset for directed modulation of micro-organism phenotype for bioprocesses and presents a comprehensive dataset for validating predictive in silico systems biology models (Carlson, 2007, 2009; Carlson & Taffs, 2010).
Methods
Strain and growth media
E. coli K-12 MG1655 was used for all experiments (μmax = 0.67 h− 1, batch growth M9 medium 10 g glucose l− 1). Individual aliquots of E. coli K-12 MG1655 were stored frozen at − 80 °C in 25 % glycerol. Media details were described previously (Folsom et al., 2014). Briefly, all experiments used M9 minimal medium with 2 × standard phosphate concentrations for additional buffering capacity. After the base M9 medium was autoclaved (84.5 mM Na2HPO4, 44.1 mM KH2PO4, 8.6 mM NaCl and 18.7 mM NH4Cl, pH 7.0 ± 0.05), 1 ml of magnesium sulfate solution (1 M MgSO4.7H2O; final concentration, 1 mM), 10 ml of trace element stock [0.55 g of CaCl2, 0.1 g of MnCl2.4H2O, 0.17 g of ZnCl2, 0.043 g of CuCl2.2H2O, 0.06 g of CoCl2.6H2O, 0.06 g of Na2MoO4.2H2O, 0.06 g of Fe(NH4)2(SO4)2.6H2O, 0.2 g of FeCl3.6H2O; adjusted to pH < 1 with HCl] and glucose were added per litre. For iron-limited growth at chemostat dilution rates of 0.1 and 0.2 h–1, the phosphate salt concentrations were 103.5 mM Na2HPO4 and 25.4 mM KH2PO4. A summary of the medium variations used to achieve ammonium-, iron- or glucose-limited conditions can be found in Table 1. Theoretical medium design calculations can be found in File S1 (available in the online Supplementary Material).
Table 1. Summary of M9 medium modifications used to establish ammonium-, iron- or glucose-limited E. coli chemostat growth.
Base M9 medium contained 2 × phosphate species for pH maintenance. Iron concentrations for the ammonium- and glucose-limited cultures included iron from the trace metals solution and estimates of typical iron concentrations in medium components, as measured experimentally.
| Concentration | |||
| Nutrient-limiting condition | Ammonium (mM) | Iron (μM) | Glucose (mM) |
| Ammonium-limited | 1.31 | 11 | 27.8 |
| Iron-limited | 18.7 | 0.022 | 27.8 |
| Glucose-limited | 18.7 | 11 | 2.2 |
Chemostat operation
Chemostats were operated as described previously (Folsom et al., 2014). Briefly, chemostats with 300 ml of medium were inoculated with 450 μl of E. coli K-12 MG1655 freezer stock and incubated without medium flow for at least 6 h at 37 °C until the reactors were turbid. The reactors were operated at four dilution rates by controlling medium volumetric feed rate. Reactors were sparged with filter-sterilized air at 1 l min− 1 and stirred at 250 r.p.m. Ambient air was humidified by preliminary sparge through sterile 37 °C nanopure water to minimize both reactor content evaporation and the associated cooling. Chemostats were operated until cultures reached steady state, defined as at least three OD600 measurements varying by around ± 10 % or less during at least two or three consecutive residence times (1/dilution rate). The different nutrient limitations and different dilution rates took a varying number of residence times to achieve steady state, ranging from six to 17 residence times. At least three separate chemostats were grown to steady state and each chemostat was sampled at least three times over two or three residence times for each combination of nutrient limitation and dilution rate. Biomass samples for proteomics analysis were harvested from reactors at the conclusion of each experiment.
Nutrient-limited medium compositions were developed which resulted in steady-state biomass OD600 values of 0.3–0.4 (Table 1). Low cell densities were selected to avoid oxygen transfer limitation (Nanchen et al., 2006). Cultures were designed to be limited for a single resource; achievement of single-nutrient limitation was supported via experimental and theoretical considerations. For ammonium-limited cultures, the chemostat effluent contained no detectable ammonium; the effluent had measurable glucose and iron. The iron-limited culture effluents had no detectable iron, but had residuals of glucose and ammonium, whilst glucose-limited culture effluents had no detectable glucose, but had detectable iron and ammonium residuals (Folsom et al., 2014). Theoretical medium calculations that supported experimental data suggesting a single nutrient limitation can be found in File S1.
Culture analysis
A correlation between OD600 and cell dry weight (CDW) was measured for ammonium-limited E. coli cultures using seven separate chemostat cultures. A coefficient of 0.38 ± 0.01 g CDW l− 1 per OD600 unit was measured. Coefficients of 0.48 ± 0.02 and 0.50 ± 0.03 g CDW l− 1 per OD600 unit were measured for iron- and glucose-limited E. coli cultures, respectively (Folsom et al., 2014). Concentrations of glucose and organic byproducts were determined using an Agilent 1200 series HPLC system equipped with an Aminex HPX-87H column (Bio-Rad). Ammonium levels were analysed using the Berthelot reaction (Searle, 1984; Stieg, 2005), adjusted to a microplate format. Biomass glycogen content was measured by a published method (Del Don et al., 1994) with modifications (File S2). Culture iron concentrations were measured using a ferene-based method as described previously (Folsom et al., 2014).
Proteomics analysis was performed as described previously (Folsom et al., 2014). Briefly, cytoplasmic and membrane protein samples were labelled with ZDYEs (ZDYE) and separated using 2D gel electrophoresis. ZDYE-stained protein spot volumes were measured using a Typhoon Trio gel scanner (GE Healthcare Life Sciences). For protein identification, blue silver-stained protein gel spots were manually excised and proteins identified using an Agilent 6520 Accurate-Mass Q-TOF Chip LC/MS system (Agilent Technologies) (Dratz & Grieco, 2009; Shipman et al., 2012).
Biomass elemental stoichiometry was determined by an external laboratory (Micro Analysis). Briefly, 150–300 ml of culture comprising 13–42 mg of CDW was washed three times with cell wash (10 mM Tris and 5 mM magnesium acetate, pH 8.0) and stored frozen ( − 80 °C). Frozen samples were thawed, pelleted and all residual liquid removed, followed by resuspension in nanopure water. The biomass-containing solution was dried at 96–100 °C for 24 h. Results of the biomass elemental analysis including Y x/N calculations and biomass glycogen measurements were analysed by ANOVA using general linear models and Tukey's method for multiple comparisons with 95 % confidence (Minitab 16).
Results
Biomass yields and specific glucose uptake rates
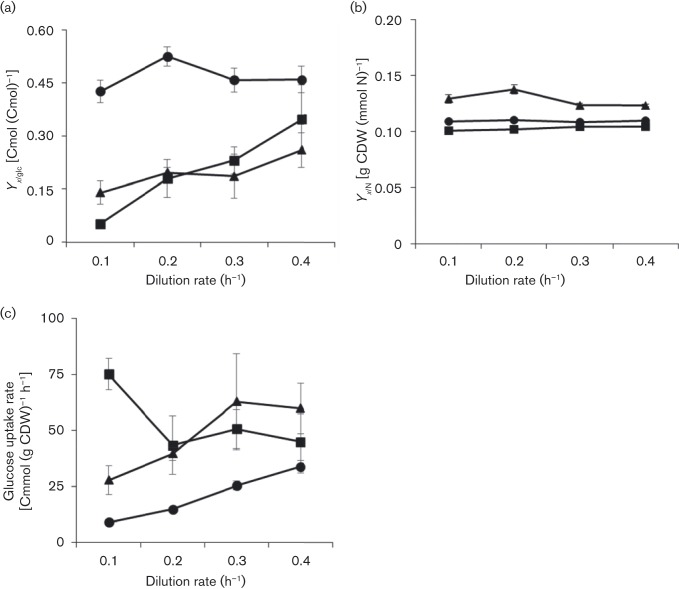
Biomass yields on glucose were a strong function of both nutrient limitation and dilution rate (D) (Fig. 1a). The biomass yield on glucose for the ammonium-limited cultures decreased around twofold from 0.26 to 0.14 Cmol biomass (Cmol glucose)–1 when the dilution rate was reduced from 0.4 to 0.1 h− 1. Biomass yields for the previously described iron-limited cultures also declined with reduced dilution rate, decreasing around sevenfold from 0.35 to 0.052 Cmol biomass (Cmol glucose)–1 when the dilution rate was reduced from 0.4 to 0.1 h− 1 (Folsom et al., 2014). Previously described glucose-limited cultures had a relatively constant Cmol biomass (Cmol glucose)–1 yield as a function of dilution rate, and the yields were higher at all dilution rates as compared with ammonium- and iron-limited cultures (Folsom et al., 2014).
Fig. 1. (a) E. coli biomass yield on glucose (Y x/glc) as a function of chemostat dilution rate and limiting nutrient (n>3). (b) Biomass per nitrogen yield (Y x/N). (n = 3, except n = 2 iron D = 0.3 h− 1 and n = 1 ammonium D = 0.1 h− 1). (c) Steady-state specific glucose uptake rate [Cmmol (g CDW)− 1 h− 1] (n>3). •, Glucose-limited; ▪, iron-limited; ▴, ammonium-limited. Data represent mean ± sem. Iron- and glucose-limited data from Folsom et al. (2014).
Biomass per nitrogen yields were a function of nutrient limitation (Fig. 1b). The ammonium-limited cultures had significantly higher biomass per nitrogen yields [g CDW (mmol N)–1] than the iron- and glucose-limited cultures (P < 0.0004, Tukey's 95 %), and the yields appeared to increase slightly with decreasing dilution rate (P = 0.083). The iron- and glucose-limited cultures had very similar biomass per nitrogen yields.
Specific glucose uptake rates (q s) were calculated using the dilution rate and measured biomass yield on glucose (q s = D/Y x/s), and were compared as a function of nutrient limitation and dilution rate (Fig. 1c). The ammonium-limited cultures had a higher specific glucose consumption rate than the glucose-limited cultures at all tested dilution rates, as expected from the lower biomass per glucose yields. Both the ammonium-limited cultures and glucose-limited cultures showed a decrease in specific glucose uptake rate with decreasing dilution rate. This was in contrast to the iron-limited cultures which exhibited such a precipitous decrease in biomass yield on glucose with decreasing dilution rate that the specific glucose uptake rate increased with decreasing dilution rate, as noted in Folsom et al. (2014). The specific glucose uptake rate for iron-limited cultures increased 1.7-fold from D = 0.4 to 0.1 h− 1. At D = 0.1 h− 1, the iron-limited culture had a specific glucose consumption rate around threefold faster than the ammonium-limited cultures and around ninefold faster than the glucose-limited cultures.
Metabolic byproduct profiles
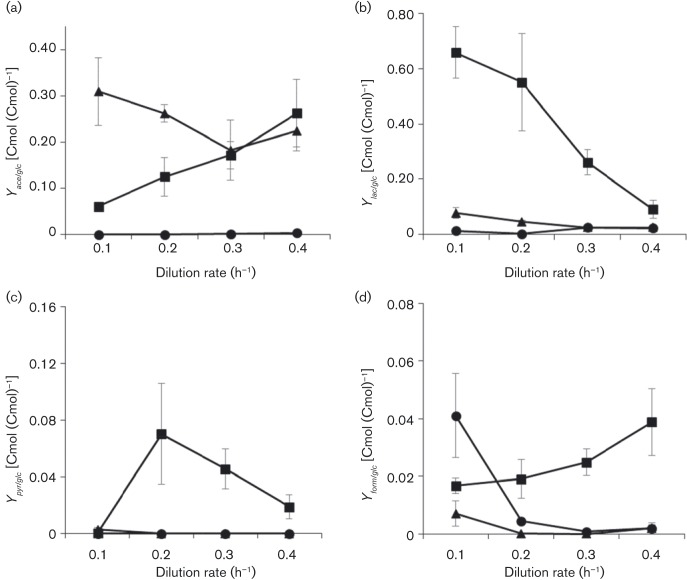
All cultures were analysed for the byproducts acetate, lactate, pyruvate and formate (Fig. 2). Cultures were also analysed for citrate, ethanol and succinate; only succinate was detected. The succinate yields were low ( < 1–2 % of metabolized glucose carbon) and can be found in the supplementary material (File S3). The absence of ethanol signal could be due to the cultures not secreting ethanol or evaporative loss in off gas. All three nutrient-limited conditions demonstrated an overflow metabolism for at least one dilution rate; here, an overflow metabolism was defined as the secretion of organic byproducts in the presence of external electron acceptor. Collectively, the byproduct profiles from the ammonium-limited cultures were markedly different from both iron- and glucose-limited cultures (Fig. 2). Ammonium-limited cultures secreted different proportions of byproducts, with acetate being the primary organic byproduct, and with lesser amounts of lactate, formate and succinate. Approximately 20 % of the metabolized glucose carbon was secreted as acetate at D = 0.4 h− 1; the percentage increased to ∼30 % at D = 0.1 h− 1. For comparison, the iron-limited cultures secreted primarily acetate (∼20 % of metabolized glucose carbon) at D = 0.4 h− 1, but acclimated to increased nutrient-limitation stress at D = 0.1 h− 1 by secreting ∼70 % of the metabolized glucose carbon as lactate (Folsom et al., 2014). The ammonium- and glucose-limited cultures secreted small but measurable amounts of formate at D = 0.1 h− 1, up to 4 % of metabolized glucose carbon (Fig. 2d). The iron-limited culture secreted measurable formate at all tested dilution rates. A summary of all data can be found in the supplementary material, including carbon and electron balances. For instance, the measured biomass and byproducts accounted for ∼80 % of the iron-limited culture carbon and electrons at D = 0.1 h− 1 (File S3). In addition, a survey of published rates and yields from ammonium-, iron- or glucose-limited chemostat growth can be found in File S4.
Fig. 2. E. coli culture organic byproduct carbon yields as a function of dilution rate and limiting nutrient: (a) acetic acid, (b) lactic acid, (c) pyruvic acid and (d) formic acid. •, Glucose-limited; ▪, iron-limited; ▴, ammonium-limited. Data represent mean ± sem (n>3). Iron- and glucose-limited data from Folsom et al. (2014).
Biomass elemental stoichiometry
Biomass carbon, hydrogen, nitrogen and oxygen elemental composition was measured for each nutrient limitation at each tested dilution rate (Table 2). The type of nutrient limitation significantly affected carbon (P = 0.04), nitrogen (P < 0.0004) and oxygen (P = 0.006) elemental composition, but not hydrogen (P>0.35) elemental composition. The experimental resolution did not measure a statistically significant difference based on dilution rate. The mean carbon content (mass basis) for the ammonium-limited E. coli cultures was 45.5 ± 0.4 %, which was significantly lower than the glucose-limited cultures (47.4 ± 0.7 %, Tukey's 95 %). The mean ammonium-limited culture biomass nitrogen content (mass basis) across the tested dilution rates was 10.8 ± 0.1 %, which was significantly lower than the biomass nitrogen contents for either iron- or glucose-limited cultures (13.3 ± 0.1 and 12.7 ± 0.2 %, respectively; Tukey's 95 %). There was no statistically significant difference between the biomass nitrogen content for the iron- and glucose-limited cultures (Tukey's 95 %). Data for the biomass oxygen and hydrogen content can be found in Table 2.
Table 2. E. coli biomass elemental stoichiometry (C, H, N and O) and mass fraction for chemostat cultures at D = 0.4, 0.3, 0.2 or 0.1 h− 1, under ammonium-, iron- or glucose-limited chemostat growth, and cellular glycogen content for D = 0.1 h− 1 .
The balance of the cellular mass was assumed to be phosphorus, sulfur and ash. Degree of reduction calculated using N2 basis. Presented means are arithmetic, unless otherwise indicated. nd, Not determined.
| D (h− 1) | n | Elemental formula | Degree of reduction | C [% ( ± sem)]* | H [% ( ± sem)]* | N [% ( ± sem)]* | O [% ( ± sem)]* | C : H | C : N | Glycogen (%)*† | |
| Ammonium-limited | |||||||||||
| 0.4 | 2 | CH1.74N0.21O0.51 | 4.71 ± | 45.53 ± | 6.62 ± | 11.28 ± | 31.29 ± | 0.57 | 4.71 | nd | |
| 0.07 | 0.80 | 0.10 | 0.34 | 2.47 | |||||||
| 0.3 | 2 | CH1.74N0.21O0.50 | 4.74 ± | 46.73 ± | 6.76 ± | 11.39 ± | 30.86 ± | 0.58 | 4.79 | nd | |
| 0.01 | 0.54 | 0.02 | 0.18 | 0.39 | |||||||
| 0.2 | 2 | CH1.69N0.20O0.58 | 4.53 ± | 44.22 ± | 6.23 ± | 10.12 ± | 34.37 ± | 0.59 | 5.10 | nd | |
| 0.08 | 0.42 | 0.30 | 0.29 | 4.49 | |||||||
| 0.1 | 3 | CH1.75N0.19O0.46 | 4.84 ± | 45.66 ± | 6.67 ± | 10.3 ± | 27.78 ± | 0.57 | 5.17 | 18.2 ± | |
| 0.10 | 0.76 | 0.24 | 0.27 | 1.50 | 0.5% | ||||||
| Mean† | 4.71 ± | 45.53 ± | 6.57 ± | 10.77 ± | 31.07 ± | ||||||
| 0.04 | 0.36 | 0.11 | 0.14 | 1.2 | |||||||
| Iron-limited | |||||||||||
| 0.4 | 2 | CH1.71N0.24O0.43 | 4.85 ± | 47.45 ± | 6.78 ± | 13.35 ± | 27.28 ± | 0.58 | 4.15 | nd | |
| 0.05 | 0.42 | 0.11 | 0.01 | 0.89 | |||||||
| 0.3 | 1 | CH1.69N0.24O0.43 | 4.83 ± | 46.07 ± | 6.47 ± | 12.92 ± | 26.46 ± | 0.59 | 4.16 | nd | |
| – | – | – | – | ||||||||
| 0.2 | 2 | CH1.70N0.24O0.40 | 4.9 ± | 47.01 ± | 6.65 ± | 13.26 ± | 25.13 ± | 0.59 | 4.14 | nd | |
| 0.05 | 0.11 | 0.04 | 0.07 | 1.41 | |||||||
| 0.1 | 4 | CH1.71N0.24O0.37 | 4.96 ± | 47.9 ± | 6.83 ± | 13.46 ± | 23.81 ± | 0.58 | 4.15 | 2.7 ± | |
| 0.06 | 0.37 | 0.08 | 0.06 | 1.86 | 0.3% | ||||||
| Mean† | 4.88 ± | 47.12 ± | 6.68 ± | 13.25 ± | 25.67 ± | ||||||
| 0.04 | 0.24 | 0.05 | 0.04 | 1.2 | |||||||
| Glucose-limited | |||||||||||
| 0.4 | 2 | CH1.74N0.23O0.45 | 4.84 ± | 47.52 ± | 6.89 ± | 12.82 ± | 28.58 ± | 0.58 | 4.33 | nd | |
| 0.10 | 0.17 | 0.06 | 0.12 | 0.57 | |||||||
| 0.3 | 2 | CH1.73N0.23O0.42 | 4.88 ± | 47.69 ± | 6.87 ± | 12.74 ± | 26.93 ± | 0.58 | 4.37 | nd | |
| 0.04 | 0.61 | 0.06 | 0.29 | 1.23 | |||||||
| 0.2 | 2 | CH1.69N0.23O0.43 | 4.82 ± | 47.26 ± | 6.67 ± | 12.49 ± | 27.35 ± | 0.59 | 4.42 | nd | |
| 0.01 | 2.19 | 0.4 | 0.59 | 1.63 | |||||||
| 0.1 | 3 | CH1.74N0.23O0.43 | 4.88 ± | 47.06 ± | 6.84 ± | 12.61 ± | 26.97 ± | 0.57 | 4.35 | 3.6 ± | |
| 0.06 | 1.5 | 0.36 | 0.45 | 0.72 | 0.3% | ||||||
| Mean† | 4.86 ± | 47.38 ± | 6.81 ± | 12.67 ± | 27.46 ± | ||||||
| 0.03 | 0.74 | 0.16 | 0.22 | 0.53 | |||||||
* Mass per cent, n = 3.
† Mean ± sem from ANOVA (least-squares).
Biomass degree of reduction was calculated using an N2 basis; a higher degree of reduction corresponded with a higher biomass energetic content, i.e. more available electrons per Cmol biomass (Roels, 1980). The iron- and glucose-limited cultures had higher mean degrees of reduction (4.88 ± 0.04 and 4.86 ± 0.03, respectively) compared with the ammonium-limited cultures (4.71 ± 0.04) (P = 0.006). Dilution rate had no significant effect on biomass degree of reduction (P = 0.08). Elemental data and a literature review can be found in File S5.
Cellular glycogen content differed between nutrient-limited conditions (Table 2). Ammonium-limited cells grown at D = 0.1 h− 1 had a glycogen content of 18.2 ± 0.5 % (mass basis) similar to other published values (Holme et al., 1957; Hua et al., 2003). Both the iron- and glucose-limited cultures had significantly lower glycogen contents when grown at D = 0.1 h− 1 (2.7 ± 0.3 and 3.6 ± 0.3 %, respectively).
Central metabolism protein abundance patterns
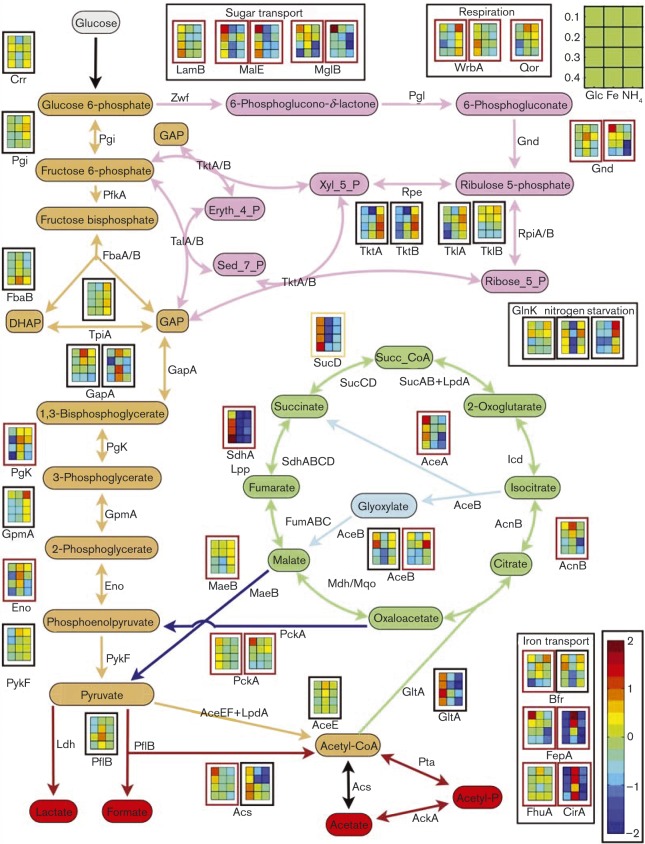
ZDYE-labelled cytosolic and membrane-associated proteins were separated by 2D gel electrophoresis; protein abundance data (Progenesis normalized spot volumes) were collected in triplicate as a function of nutrient limitation and dilution rate (representative ZDYE-labelled protein gel images can be found in File S6). Abundance profiles for proteins associated with the central metabolism and limiting-nutrient transporters are presented in Fig. 3. Many proteins were discovered in multiple gel spots, suggesting multiple isoforms which were likely the result of protein post-translational modifications (Folsom et al., 2014); these isoform data highlighted a major advantage of gel-based proteomics techniques over gel-free techniques (Chevalier, 2010). To simplify presentation and interpretation of abundance data in Fig. 3, proteins found in multiple gel spots were summed if the individual abundance patterns had a Pearson distance metric ≤ 0.45 (heuristic); otherwise, the data were presented as separate abundance trends. Individual protein spot data can be found in Files S7–S12.
Fig. 3. E. coli protein abundance profiles for identified central metabolism proteins and transporter proteins. Key for nutrient-limited conditions is located in the upper right of figure, briefly dilution rate (h− 1) is on the y-axis [high stress on top (0.1 h− 1), low stress on bottom (0.4 h− 1)] and nutrient limitation on the x-axis (glucose, iron or ammonium limitation from left to right). Red box outlines correspond with proteins found in the membrane fraction, black box outlines correspond with proteins found in the cytoplasm fraction, and tan box outlines represent aggregate protein isoforms found in both the membrane and cytoplasm fractions. Many proteins had multiple isoforms; when multiple protein isoforms had a Pearson distance ≤ 0.45, they were presented as a summed trend. Colour bar indicates fold change ( < –2 to >2) compared with mean spot volume (log2 scale). Data for all individual proteins can be found in Files S7–S12. Iron- and glucose-limited data from Folsom et al. (2014). GAP, glyceraldehyde 3-phosphate; Xyl_5_P, xylulose-5-phosphate; Eryth_4_P, erythrose-4-phosphate; Sed_7_P, sedoheptulose-7-phosphate; ribose_5_P, ribose 5-phosphate; DHAP, dihydroxyacetone phosphate; Succ_CoA, succinyl-CoA.
Generally, abundance levels for glycolysis-associated enzymes were higher for the ammonium- and iron-limited cultures compared with the glucose-limited cultures. For example, phosphoglucose isomerase (Pgi), triose phosphate isomerase (TpiA) and glycerate phosphoglycerate mutase (GpmA) abundances were highest during ammonium-limited growth whilst fructose bisphosphate aldolase (FbaB), glyceraldehyde 3-phosphate dehydrogenase (GapA), phosphoglycerate kinase (Pgk) and enolase (Eno) abundances were highest during iron-limited growth (Fig. 3). These trends were consistent with the higher specific glucose uptake rates measured for the respective cultures compared with glucose-limited cultures. Abundance of the two pyruvate formate lyase isoforms (PflB, ID 0200, eID 0504) was also higher under iron-limited conditions compared with the other tested conditions.
Ammonium-limited growth had a high relative protein abundance for some pentose phosphate pathway (PPP) enzymes, including transketolase (TktA, TktB) and transaldolase (TalA, TalB), compared with iron- or glucose-limited growth. However, abundance of 6-phosphogluconate dehydrogenase (Gnd, decarboxylating enzyme from the oxidative portion of the PPP) isoforms (eID 0382, eID 0383) was low during ammonium-limited growth compared with the other conditions. Gnd isoform eID 0383 had a low abundance for both ammonium- and glucose-limited growth. Low abundance of this enzyme probably limited flux through the oxidative PPP branch or potentially directed gluconate 6-phosphate flux toward the Entner–Doudorouff pathway.
Ammonium- and iron-limited cultures had a low abundance of key tricarboxylic acid (TCA) cycle enzymes, including citrate synthase (GltA), isocitrate lyase (AceA), succinyl-CoA synthetase (SucD) and succinate dehydrogenase (SdhA, gel spot also contained Lpp protein signal), compared with glucose-limited cultures, likely indicative of reduced TCA fluxes. Aconitase (AcnB) abundance was a departure from this trend. During iron-limited growth, AcnB abundance was up to 2.4-fold higher than during ammonium- or glucose-limited growth. A discussion of AcnA and AcnB abundance and how they relate to iron-limited growth can be found in Folsom et al. (2014). Acetyl-CoA synthetase (Acs) and anaplerotic reaction phosphoenolpyruvate carboxykinase (PckA) had a higher abundance during glucose-limited growth relative to ammonium- or iron-limited growth. The glucose-limited cultures demonstrated an increase in abundance of glyoxylate shunt enzymes AceA and AceB as a function of reduced dilution rate, a trend consistent with previous fluxomic studies (Fischer & Sauer, 2003).
Glutamate decarboxylase (GadAB isoforms eID 0742 and eID 0915), an enzyme associated with acclimation to acid stress (Moreau, 2007), had a higher abundance during both ammonium- and iron-limited conditions relative to glucose-limited growth. The trend was consistent with the elevated levels of secreted organic acids although seven isoforms of GadAB were identified with five occurring in spots with multiple proteins making conclusive interpretations difficult (Files S7–S12).
Two enzymes from the electron transport chain were identified during proteome analysis: alternative quinone oxidoreductases WrbA and Qor. WrbA was found as three isoforms, whilst Qor was identified in one spot. Membrane WrbA (eID 0051) was present at higher abundance during glucose-limited growth compared with ammonium- and iron-limited growth, whilst the cytosolic WrbA isoform (eID 0443) was synthesized at higher levels during ammonium-limited growth for unknown reasons.
Transporter and porin protein expression
Enzymes associated with acquisition of the limiting nutrient had generally higher abundances. Examples include the iron-processing proteins FepA and CirA during iron-limited growth, and the sugar transport-associated MalE and MglB proteins during glucose-limited growth (Fig. 3) (Folsom et al., 2014). Proteomic analysis identified a large number of porin and other transporter proteins (e.g. OppA, OmpX, OmpT, OmpF, OmpA and TolC). Abundance patterns were diverse and many of the proteins such as OmpA had many isoforms. OmpA was identified in 18 separate protein spots, sometimes as the sole protein and sometimes along with other proteins. The OmpA protein isoforms demonstrated many different trends across nutrient limitation and dilution rate; however, the trends were beyond current knowledge to interpret. Porin and transporter protein data can be found in Files S7–S12.
Discussion
Ecological and economic theories have been used to predict and interpret tradeoffs between resource allocation and cellular function (Bloom et al., 1985; Carlson & Taffs, 2010; Miller et al., 2005; Molenaar et al., 2009; Tilman, 1982). These tradeoffs are thought to influence the observations in the current chemostat study where E. coli is constrained by the availability of a single nutrient. Investing the scarce nutrient into one function precludes it from being invested into other functions, preventing the metabolism from optimizing the use of all nutrients simultaneously and resulting in overflow metabolisms. These results are a demonstration of the ecological null hypothesis and thought exercise known as the Darwinian Demon (Law, 1979). Superspecies which optimize all functions simultaneously, such as the Darwinian Demon, have not been observed in nature or the laboratory due to resource allocation constraints on function.
Metabolic tradeoffs resulting in an overflow metabolism were observed on two levels: (i) in the investment of resources such as nitrogen into different competitive parallel pathways and (ii) in the varied use of metabolites such as pyruvate as a metabolic intermediate, energy source or redox sink. E. coli has a flexible central metabolism with numerous parallel pathways which permit a variety of phenotypes (Carlson, 2007). Each parallel pathway requires different enzyme sets with different amino acid requirements and therefore different nutrient investment requirements. The pathways also oxidize substrates with different catabolic efficiencies. High-efficiency biochemical pathways such as the oxidative TCA cycle or the high phosphate/oxygen number respiration pathway components (e.g. Nuo) comprise large, often multi-subunit, proteins which require large resource investments to synthesize compared with fermentative strategies. Based on the measured proteome, the ammonium- and iron-limited cultures likely downregulated oxidative TCA cycle fluxes and instead used branched TCA cycle fluxes to produce essential intermediates such as 2-oxoglutarate (Clark, 1989). These acclimations are highly efficient in terms of nitrogen or iron investment. The observed overflow physiologies require fewer enzymic steps relative to a complete TCA cycle and utilize enzymes that are typically smaller than the enzymes found in the TCA cycle or in the Nuo complex (Carlson, 2007), yet maintain essential fluxes. However, these strategies come with a tradeoff: they require large glucose fluxes to offset the low ATP per glucose yields from substrate-level phosphorylation compared with oxidative phosphorylation (Fig. 1c). Use of the glyoxylate shunt by the glucose-limited cultures is also a reflection of the same metabolic tradeoff concepts (Fischer & Sauer, 2003; Folsom et al., 2014). The glucose-limited metabolism cannot simultaneously optimize anabolic and catabolic functions, resulting in metabolic tradeoffs as described previously (Carlson, 2007).
The fate of pyruvate under an iron-scarcity gradient illustrates how metabolic tradeoffs can operate on metabolite pools in addition to pathway enzymes. During nutrient-sufficient growth, pyruvate serves as both a metabolic intermediate and an electron source being oxidized to acetyl-CoA, NADH and CO2 by the pyruvate dehydrogenase complex (PDHc). Acetyl-CoA can be further oxidized in the TCA cycle and the NADH oxidized by the respiratory chain; these pathway enzymes require large anabolic resource investments to synthesize, but the pathways ultimately generate substantial ATP. During modest iron stress the cellular economics change; pyruvate is best utilized as only a substrate-level phosphorylation ATP source. It is oxidized to acetyl-CoA, using pyruvate formate lyase which avoids the formation of additional NADH (which would require further enzyme synthesis to oxidize and associated resource investment), followed by generation of ATP via phosphate acetyltransferase and acetate kinase, ultimately leading to the secretion of acetate and formate. This strategy requires no net iron investment, generates no net reducing equivalents, yet extracts energy from pyruvate. During severe iron-limitation stress (e.g. D = 0.1 h− 1), the high NADH/NAD+ ratios shift the cellular economics further so pyruvate is best utilized as an electron sink, rather than energy source, leading to lactate synthesis. Lactate production requires no iron investment and sinks 12 available electrons versus the 10 available electrons found in the combination of one acetate and one formate (Roels, 1980). Lactate synthesis serves as an efficient electron valve; however, it comes with the economic penalty of a lower ATP per glucose yield than the other two described strategies for metabolizing pyruvate.
Formate secretion was measured for ammonium-, iron- and glucose-limited cultures at low dilution rates (e.g. D = 0.1 h− 1); this is not unheard of, and formate has been measured previously in oxic and microaerophilic E. coli cultures (Alexeeva et al., 2000; Castan & Enfors, 2002; De Maeseneire et al., 2006). The primary formate synthesis route in E. coli is pyruvate formate lyase, although it is theoretically possible formate could be produced from tetrahydrofolate metabolism. The glucose-limited cultures grown at D = 0.1 h− 1 secreted formate; the cultures were agitated and sparged at the same rates as the cultures grown at D = 0.4 h− 1 which did not produce formate or acetate. Therefore, bulk-phase oxygen mass transfer limitation is not thought to be responsible for the formate secretion. Alternatively, it has been suggested that an extracellular polymer such as DNA could interfere with oxygen mass transfer at the cell membrane (Castan & Enfors, 2002). This seems unlikely here considering the cultures did not produce appreciable amounts of acetate which is typically associated with oxygen-limited E. coli growth (Alexeeva et al., 2000). The previously discussed nutrient investment versus cellular function metabolic tradeoff theory represents another possible hypothesis. Pyruvate formate lyase is a relatively small enzyme with modest nutrient requirements for synthesis compared with the large PDHc (1520 versus 42 096 aa, respectively; Carlson, 2007), yet both catalyse the conversion of pyruvate to acetyl-CoA, albeit with different coproducts. Under nutrient-limited conditions, pyruvate formate lyase may represent a better return on limiting resource than PDHc. In fact, in silico resource tradeoff analysis predicted correctly that formate would be the first reduced byproduct secreted by E. coli transitioning down a carbon investment tradeoff curve (Carlson, 2007). This prediction is not intuitive and highlights the power of in silico systems biology. However, pyruvate formate lyase is sensitive to oxygen damage (Becker et al., 1999). Operation of pyruvate formate lyase under oxic conditions might be possible if the damaged portion of the enzyme were repaired continually. YfiD is thought to replace the oxygen-damaged portion of pyruvate formate lyase (Wagner et al., 2001; Zhu et al., 2007) creating a theoretical mechanism that could permit pyruvate formate lyase to function under oxic conditions. The repair protein is small (127 aa), and a damaged protein could be replaced many times and still require fewer resources than a single PDHc. For instance, the repair protein could be synthesized, utilized, damaged and discarded >300 times, and still require fewer amino acids than one PDHc. It is unlikely that E. coli would discard the amino acids in the damaged protein and would instead recycle most of the resources, further adding to the theoretical return on resource investment. The decreasing formate per glucose trend observed in the iron-limited cultures is also consistent with the published in silico tradeoff analysis; at extreme levels of resource stress, it is predicted that flux through pyruvate formate lyase would decrease and flux through lactate dehydrogenase would increase. Interestingly, the ammonium-limited cultures only coproduced formate and acetate at D = 0.1 h− 1; metabolic tradeoff theory predicts acetate should be coproduced with formate. This discrepancy from theory will require additional research to decode.
The observed phenotypes were likely a summation of numerous, concurrent processes. In addition to the discussed metabolic tradeoff considerations, other processes such as growth rate effects, maintenance energy expenditures and population phenotype distributions could have contributed to observed phenotypes. The combined ammonium-, iron- and glucose-limited culture datasets provide a basis for comparing the effect of specific growth rate on phenotype. The ammonium-, iron- and glucose-limited cultures at D = 0.1 h− 1 demonstrated very different phenotypes whilst growing at the same specific growth rate, suggesting nutrient limitation and the associated stresses had a larger role on the phenotypes than specific growth rate alone. Maintenance energy expenditures are often evoked to explain decreases in biomass yields on electron donor at low specific growth rates. At low specific growth rates, the maintenance energy expenditures represent a larger fraction of the total cellular budget, reducing observed biomass yields (Carlson & Srienc, 2004; Neijssel et al., 1996; Pirt, 1982). Maintenance energy certainly played a role in the current study; however, the current data demonstrate that the cellular phenotype and the associated biomass per glucose yield change as a function of nutrient limitation and dilution rate. These phenotypic changes will require a re-examination of common assumptions used to calculate maintenance energy (research in progress). The role of population phenotype distributions on the measured properties is unknown but likely important (Kussell & Leibler, 2005; Natarajan & Srienc, 2000). The different nutrient limitations and dilution rates resulted in different stress severities as reflected in the biomass per glucose yields and likely reflected in the unmeasured RpoS levels (Ihssen & Egli, 2004). The stress could have influenced the distribution of cellular phenotypes, including the distribution of active and inactive ‘persistor’ cells (Kussell et al., 2005). These cellular distributions are difficult to measure and unfortunately were not measured here.
Biomass per iron yields are challenging to measure due to the low aqueous iron solubility under oxic culturing conditions, the tendency of Fe(III) to form high-molecular-mass polymers (Wang & Newton, 1969), small cellular iron content, complexities distinguishing between physiologically relevant intracellular iron and non-specific, extracellularly bound iron, and the influence of contaminating iron sources, such as tap water, medium reagents and laboratory equipment. Our work calculated biomass per iron yields of 100 000–150 000 g CDW (g Fe)− 1 during steady state, iron-limited chemostat growth on defined medium (Folsom et al. 2014). Contaminating iron from exogenous sources prevented measurement of biomass per iron yields during ammonium- or glucose-limited growth experiments; however, secreted, steady-state siderophore levels, as represented by chromazurol-S activity, were measured for all tested conditions (File S3). Biomass per iron yields of 5000–7000 g CDW (g Fe)− 1 have been reported during batch cultivation on both defined and complex medium under well-established, non-iron-limited conditions (Hudson et al. 1993; Abdul-Tehrani et al. 1999). The same researchers reported biomass yields of ∼50 000 g CDW (g Fe)− 1 during batch cultivation in iron-limited, defined medium (Abdul-Tehrani et al. 1999). These yields are within the range (two- to threefold) of the values reported in our studies. The modest differences in measured yields could be due to steady-state, chemostat growth conditions versus unsteady-state, batch cultivation conditions or the sensitivity of the biomass per iron yield to small experimental variations. The often cited bacterial biomass per iron yield of 200 g CDW (g Fe)− 1 (Pirt, 1975) differs substantially, for unknown reasons, from the studies mentioned here.
The current study presents a comprehensive dataset detailing acclimations to common nutrient-limitation stresses. The observed phenotypes are believed to reflect strategic investments of the limiting resource into essential metabolic pathways which result in suboptimal use of some non-limiting resources; the tradeoff economics were nutrient type and chemostat dilution rate dependent. The data provide additional insight into evolutionary selection and provide data for designing bioprocesses and for testing predictive in silico systems biology models.
Acknowledgements
The authors would like to thank Reed Taffs for technical assistance and helpful discussions, Jonathan Hilmer, Walid Maaty and the Montana State University Mass Spectrometry, Proteomics & Metabolomics Facility for technical support, Matthew Shipman and Mark Burr for protocol assistance, Ashley Beck and Kristopher Hunt for careful manuscript reading, Albert Parker for statistics assistance, and ZDYE LLC for the generous gift of ZDYEs. The authors would also like to thank two anonymous reviewers for their helpful suggestions. This work was funded by the National Institutes of Health (P20RR024237) and the National Science Foundation Integrative Graduate Education and Research Traineeship program (DGE 0654336). The Murdock Charitable Trust provided generous support for the instrumentation in the Mass Spectrometry Facility.
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Abbreviations:
- CDW
cell dry weight
- PDHc
pyruvate dehydrogenase complex
- PPP
pentose phosphate pathway
- TCA
tricarboxylic acid
References
- Abdul-Tehrani H., Hudson A.J., Chang Y.S., Timms A.R., Hawkins C., Williams J.M., Harrison P.M., Guest J.R., Andrews S.C. (1999). Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient J Bacteriol 181 1415–1428 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeeva S., de Kort B., Sawers G., Hellingwerf K.J., de Mattos M.J.T. (2000). Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli J Bacteriol 182 4934–4940 10.1128/JB.182.17.4934-4940.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S., Ziv E. (1976). Dense growth of aerobic bacteria in a bench-scale fermentor Biotechnol Bioeng 18 81–94 10.1002/bit.260180107 . [DOI] [PubMed] [Google Scholar]
- Becker A., Fritz-Wolf K., Kabsch W., Knappe J., Schultz S., Volker Wagner A.F. (1999). Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase Nat Struct Biol 6 969–975 10.1038/13341 . [DOI] [PubMed] [Google Scholar]
- Bloom A.J., Chapin F.S., Mooney H.A. (1985). Resource limitation in plants – an economic analogy Annu Rev Ecol Evol Syst 16 363–392 10.1146/annurev.es.16.110185.002051. [DOI] [Google Scholar]
- Brauer M.J., Yuan J., Bennett B.D., Lu W., Kimball E., Botstein D., Rabinowitz J.D. (2006). Conservation of the metabolomic response to starvation across two divergent microbes Proc Natl Acad Sci U S A 103 19302–19307 10.1073/pnas.0609508103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull A.T. (2010). The renaissance of continuous culture in the post-genomics age J Ind Microbiol Biotechnol 37 993–1021 10.1007/s10295-010-0816-4 . [DOI] [PubMed] [Google Scholar]
- Carlson R.P. (2007). Metabolic systems cost-benefit analysis for interpreting network structure and regulation Bioinformatics 23 1258–1264 10.1093/bioinformatics/btm082 . [DOI] [PubMed] [Google Scholar]
- Carlson R.P. (2009). Decomposition of complex microbial behaviors into resource-based stress responses Bioinformatics 25 90–97 10.1093/bioinformatics/btn589 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson R., Srienc F. (2004). Fundamental Escherichia coli biochemical pathways for biomass and energy production: creation of overall flux states Biotechnol Bioeng 86 149–162 10.1002/bit.20044 . [DOI] [PubMed] [Google Scholar]
- Carlson R.P., Taffs R.L. (2010). Molecular-level tradeoffs and metabolic adaptation to simultaneous stressors Curr Opin Biotechnol 21 670–676 10.1016/j.copbio.2010.05.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castan A., Enfors S.O. (2002). Formate accumulation due to DNA release in aerobic cultivations of Escherichia coli Biotechnol Bioeng 77 324–328 10.1002/bit.1198 . [DOI] [PubMed] [Google Scholar]
- Chevalier F. (2010). Highlights on the capacities of “gel-based” proteomics Proteome Sci 8 23 10.1186/1477-5956-8-23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowski T.H., Grover J.P. (2008). Element content of Pseudomonas fluorescens varies with growth rate and temperature: a replicated chemostat study addressing ecological stoichiometry Limnol Oceanogr 53 1242–1251 10.4319/lo.2008.53.4.1242. [DOI] [Google Scholar]
- Clark D.P. (1989). The fermentation pathways of Escherichia coli FEMS Microbiol Rev 5 223–234 . [DOI] [PubMed] [Google Scholar]
- Cordier J.-L., Butsch B., Birou B., Stockar U. (1987). The relationship between elemental composition and heat of combustion of microbial biomass Appl Microbiol Biotechnol 25 305–312. [Google Scholar]
- Cotner J.B., Makino W., Biddanda B.A. (2006). Temperature affects stoichiometry and biochemical composition of Escherichia coli Microb Ecol 52 26–33 10.1007/s00248-006-9040-1 . [DOI] [PubMed] [Google Scholar]
- De Maeseneire S.L., De Mey M., Vandedrinck S., Vandamme E.J. (2006). Metabolic characterisation of E. coli citrate synthase and phosphoenolpyruvate carboxylase mutants in aerobic cultures Biotechnol Lett 28 1945–1953 10.1007/s10529-006-9182-8 . [DOI] [PubMed] [Google Scholar]
- Del Don C., Hanselmann K.W., Peduzzi R., Bachofen R. (1994). Biomass composition and methods for the determination of metabolic reserve polymers in phototrophic sulfur bacteria Aquat Sci 56 1–15 10.1007/BF00877431. [DOI] [Google Scholar]
- Dratz E.A., Grieco P. (2009). Novel optical labeling molecules in proteomics and other biological analyses World Patent 2009/005871-A. [Google Scholar]
- Egli T. (1991). On multiple-nutrient-limited growth of microorganisms, with special reference to dual limitation by carbon and nitrogen substrates Antonie van Leeuwenhoek 60 225–234 10.1007/BF00430367 . [DOI] [PubMed] [Google Scholar]
- Fischer E., Sauer U. (2003). A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli J Biol Chem 278 46446–46451 10.1074/jbc.M307968200 . [DOI] [PubMed] [Google Scholar]
- Folsom J.P., Parker A.E., Carlson R.P. (2014). Physiological and proteomic analysis of Escherichia coli iron-limited chemostat growth J Bacteriol 196 2748–2761 10.1128/JB.01606-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldal M., Norland S., Fagerbakke K., Thingstad F., Bratbak G. (1996). The elemental composition of bacteria: a signature of growth conditions? Mar Pollut Bull 33 3–9 10.1016/S0025-326X(97)00007-6. [DOI] [Google Scholar]
- Ho K.P., Payne W.J. (1979). Assimilation efficiency and energy contents of prototrophic bacteria Biotechnol Bioeng 21 787–802 10.1002/bit.260210505. [DOI] [Google Scholar]
- Holme T., Westöö G., Svennerholm L., Magnéli A., Magnéli A., Pestmalis H., Åsbrink S. (1957). Continuous culture studies on glycogen synthesis in Escherichia coli B Acta Chem Scand 11 763–775 10.3891/acta.chem.scand.11-0763. [DOI] [Google Scholar]
- Hua Q., Yang C., Baba T., Mori H., Shimizu K. (2003). Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts J Bacteriol 185 7053–7067 10.1128/JB.185.24.7053-7067.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q., Yang C., Oshima T., Mori H., Shimizu K. (2004). Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures Appl Environ Microbiol 70 2354–2366 10.1128/AEM.70.4.2354-2366.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A.J., Andrews S.C., Hawkins C., Williams J.M., Izuhara M., Meldrum F.C., Mann S., Harrison P.M., Guest J.R. (1993). Overproduction, purification and characterization of the Escherichia coli ferritin Eur J Biochem 218 985–995 10.1111/j.1432-1033.1993.tb18457.x . [DOI] [PubMed] [Google Scholar]
- Ihssen J., Egli T. (2004). Specific growth rate and not cell density controls the general stress response in Escherichia coli Microbiology 150 1637–1648 10.1099/mic.0.26849-0 . [DOI] [PubMed] [Google Scholar]
- Keshavarz T., Roy I. (2010). Polyhydroxyalkanoates: bioplastics with a green agenda Curr Opin Microbiol 13 321–326 10.1016/j.mib.2010.02.006 . [DOI] [PubMed] [Google Scholar]
- Kussell E., Leibler S. (2005). Phenotypic diversity, population growth, and information in fluctuating environments Science 309 2075–2078 10.1126/science.1114383 . [DOI] [PubMed] [Google Scholar]
- Kussell E., Kishony R., Balaban N.Q., Leibler S. (2005). Bacterial persistence: a model of survival in changing environments Genetics 169 1807–1814 10.1534/genetics.104.035352 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law R. (1979). Optimal life histories under age-specific predation Am Nat 114 399–417 10.1086/283488. [DOI] [Google Scholar]
- Liu X., Ferenci T. (1998). Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli J Bacteriol 180 3917–3922 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T.E., Burns J.H., Munguia P., Walters E.L., Kneitel J.M., Richards P.M., Mouquet N., Buckley H.L. (2005). A critical review of twenty years’ use of the resource-ratio theory Am Nat 165 439–448 10.1086/428681 . [DOI] [PubMed] [Google Scholar]
- Molenaar D., vanBerlo R., deRidder D., Teusink B. (2009). Shifts in growth strategies reflect tradeoffs in cellular economics Mol Syst Biol 5 323 10.1038/msb.2009.82 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J. (1950). [The continuous culture technique: theory and applications] Ann Inst Pasteur (Paris) 79 390 (in French). [Google Scholar]
- Moreau P.L. (2007). The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids J Bacteriol 189 2249–2261 10.1128/JB.01306-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanchen A., Schicker A., Sauer U. (2006). Nonlinear dependency of intracellular fluxes on growth rate in miniaturized continuous cultures of Escherichia coli Appl Environ Microbiol 72 1164–1172 10.1128/AEM.72.2.1164-1172.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A., Srienc F. (2000). Glucose uptake rates of single E. coli cells grown in glucose-limited chemostat cultures J Microbiol Methods 42 87–96 10.1016/S0167-7012(00)00180-9 . [DOI] [PubMed] [Google Scholar]
- Neijssel O.M., Teixeira de Mattos M.J., Tempest D.W. (1996). Growth yield and energy distribution. In Escherichia coli and Salmonella: Cellular and Molecular Biology, pp. 1683–1692. Edited by Neidhardt F. C. Washington, DC: American Society for Microbiology. [Google Scholar]
- Novick A., Szilard L. (1950). Description of the chemostat Science 112 715–716 10.1126/science.112.2920.715 . [DOI] [PubMed] [Google Scholar]
- Pickett A.M., Bazin M.J., Topiwala H.H. (1979). Growth and composition of Escherichia coli subjected to square-wave perturbations in nutrient supply: effect of varying frequencies Biotechnol Bioeng 21 1043–1055 10.1002/bit.260210609. [DOI] [Google Scholar]
- Pirt S.J. (1975). Principles of Microbe Cultivation Oxford: Blackwell Scientific. [Google Scholar]
- Pirt S.J. (1982). Maintenance energy: a general model for energy-limited and energy-sufficient growth Arch Microbiol 133 300–302 10.1007/BF00521294 . [DOI] [PubMed] [Google Scholar]
- Roels J.A. (1980). Application of macroscopic principles to microbial metabolism Biotechnol Bioeng 22 2457–2514 10.1002/bit.260221202. [DOI] [PubMed] [Google Scholar]
- Sauer U., Lasko D.R., Fiaux J., Hochuli M., Glaser R., Szyperski T., Wüthrich K., Bailey J.E. (1999). Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism J Bacteriol 181 6679–6688 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep M., Ryall B., Ferenci T. (2012). The identification of global patterns and unique signatures of proteins across 14 environments using outer membrane proteomics of bacteria Mol Biosyst 8 3017–3027 10.1039/c2mb25212k . [DOI] [PubMed] [Google Scholar]
- Scott J.T., Cotner J.B., Lapara T.M. (2012). Variable stoichiometry and homeostatic regulation of bacterial biomass elemental composition Front Microbiol 3 42 10.3389/fmicb.2012.00042 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle P.L. (1984). The Berthelot or indophenol reaction and its use in the analytical chemistry of nitrogen: a review Analyst (Lond) 109 549–568 10.1039/an9840900549. [DOI] [Google Scholar]
- Senior P.J. (1975). Regulation of nitrogen metabolism in Escherichia coli Klebsiella aerogenes: studies with the continuous-culture technique J Bacteriol 123 407–418 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman M., Lubick K., Fouchard D., Guram R., Grieco P., Jutila M., Dratz E.A. (2012). Proteomic and systems biology analysis of monocytes exposed to securinine, a GABAA receptor antagonist and immune adjuvant PLoS One 7 e41278 10.1371/journal.pone.0041278 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonds S., Grover J.P., Chrzanowski T.H. (2010). Element content of Ochromonas danica: a replicated chemostat study controlling the growth rate and temperature FEMS Microbiol Ecol 74 346–352 10.1111/j.1574-6941.2010.00963.x . [DOI] [PubMed] [Google Scholar]
- Sterner R.W., Elser J.J. (2002). Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere Princeton, NJ: Princeton University Press. [Google Scholar]
- Stieg S. (2005). 4500-NH3 F. Phenate method. In Standard Methods for the Examination of Water and Wastewater, pp. 4–114. Edited by Eaton A. D., Clesceri L. S., Rice E. W., Greenberg A. E., Franson M. A. H. Washington, DC: American Public Health Association/American Water Works Association/Water Environment Federation. [Google Scholar]
- Sukmarini L., Shimizu K. (2010). Metabolic regulation of Escherichia coli and its glnG and zwf mutants under nitrogen limitation Biochem Eng J 48 230–236 10.1006/bbrc.2001.5186 . [DOI] [PubMed] [Google Scholar]
- Tilman D. (1982). Resource Competition and Community Structure., Princeton, NJ: Princeton University Press. [PubMed] [Google Scholar]
- Wagner A.F., Schultz S., Bomke J., Pils T., Lehmann W.D., Knappe J. (2001). YfiD of Escherichia coli and Y06I of bacteriophage tbl4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase Biochem Biophys Res Commun 285 456–462 10.1006/bbrc.2001.5186 . [DOI] [PubMed] [Google Scholar]
- Wang C.C., Newton A. (1969). Iron transport in Escherichia coli: roles of energy-dependent uptake and 2,3-dihydroxybenzoylserine J Bacteriol 98 1142–1150 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Shalel-Levanon S., Bennett G., San K.Y. (2007). The YfiD protein contributes to the pyruvate formate-lyase flux in an Escherichia coli arcA mutant strain Biotechnol Bioeng 97 138–143 10.1002/bit.21219 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data