Abstract
During a collecting trip exploring new habitats and locations for keratinophilic fungi, soil samples were collected from Bohani village in the central Indian state of Madhya Pradesh. Following isolation using a hair baiting technique, one sample yielded an interesting hyphomycetous fungus. The fungus, which could easily be observed under a dissecting microscope, formed prominently large, dark brown to opaque, fusoid conidia on horse hair after 2 mo of incubation. Single-conidial colonies were cultivated on Sabouraud dextrose agar medium by direct transfer, using a fine needle under a dissecting microscope. Morphologically the fungus resembles genera such as Bahugada, Hadrosporium, Manoharachariella, Pithomyces, and Septosporiopsis, but differs with regard to its conidia and conidiogenous cells. Based on its unique morphology, the present collection is described as a new genus, Matsushimamyces, with M. bohaniensis as type species. Phylogenetic analysis of the LSU region placed Matsushimamyces in Latoruaceae (Pleosporales, Dothideomycetes). Furthermore, based on the morphological and phylogenetic similarity (ITS and LSU rDNA), a second species, Polyschema venustum, is also referred to Matsushimamyces.
Keywords: DNA phylogeny, Pithomyces, Polyschema, soil baiting, systematics
INTRODUCTION
Tropical regions of the world harbour an enormous biodiversity, including fungi (Hawksworth 1998). India is a vast country full of diverse habitats that are rich in fungal diversity, a large portion of which remains unknown. The notion that non-hotspot regions in India also harbour a large number of unknown species is supported from recent discoveries, such as those of Arthrinium jatrophae, Auxarthronopsis bandhavgarhensis, Chaetomium jatrophae, and Gymnoascus verruvosus (Sharma et al. 2013, 2014, Sharma & Singh 2013). Two biodiversity hotspots were identified in India, namely the north-eastern Himalayas and the Western Ghats (Myers et al. 2000). After this demarcation, there was a marked increase in the number of fungal diversity exploration studies in these two areas, resulting in the discovery of several new fungi. This did, however, result in a diversion of focus from other regions in India, from which novel fungi still await collection or isolation and description. The present communication is a report of one such study from a village in Central India.
MATERIAL AND METHODS
Isolates
Soil was collected in sealed polythene bags using a sterile spatula, and brought to the laboratory for processing. The soil sample was stored at room temperature until processed. A slight modification of Vanbreusegham’s hair baiting technique was employed for recovery of fungi from soil (Sharma & Rajak 2003). Stereo microscopic observations were made under a UNICON dissecting microscope (Unique Scientific Traders, Ambala, India). Single conidial colonies were obtained by removing the conidia with the help of a fine needle and plating them on Sabouraud dextrose agar medium (SDA; Hi-Media and Merck, Mumbai) and 2 % potato dextrose agar (PDA; Merck, Mumbai). Fungal-colonised horse hair was mounted in lactophenol and visualised under an Olympus BX53 (Olympus Corporation, Japan) and photographed with a ProgRes C5 camera (Jenoptik, AL) as well as under a Nikon Eclipse Ni microscope with attached D5100 camera. The ex-type culture(s) of the fungus were deposited in the Centre for Biodiversity Exploration and Conservation (CBEC), Jabalpur, Microbial Culture Collection (MCC) in Pune, and the Microbial Type Culture Collection (MTCC) in Chandigarh.
DNA isolation, phylogeny and analysis
Fungal genomic DNA was extracted following the CTAB method described by Voigt et al. (1999). The only modification was in the crushing step. The samples were crushed using a micro-pestle (GV-28541, Geovista, India) for 1–2 min. The concentrations of extracted DNAs were measured by a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA). The quality of the extracted DNA was confirmed by agarose gel electrophoresis. PCR amplification of the ITS region was carried out on Thermal cycler ABI 2720 (Applied Biosystems, CA). For amplification of the ITS1-5.8S-ITS2 region, the primer pairs ITS1 and ITS4 were used (White et al. 1990). The PCR amplified product was separated on 1.5 % agarose gel, and visualised by ethidium bromide staining. It was purified by PEG-NaCl (Polyethylene glycol-NaCl) as described by Sambrook et al. (1989). Sequencing was performed on an ABI 3730 XL DNA analyser (Applied Biosystems, CA) using the ABI Big-Dye terminator v. 3.1 cycle sequencing kit as per the manufacturer’s instructions, with sequences deposited in the National Center for Biotechnology Information (NCBI). Forward and reverse sequence reads were assembled, and the contigs were obtained using Chromas Pro software. A NCBI BLASTn search for the ITS region was carried out for sequence similarity (Altschul et al. 1990). Sequences were aligned using Clustal W of Molecular Evolutionary Genetics Analysis (MEGA v. 5.0; Tamura et al. 2011). A phylogenetic tree for the LSU region was constructed using the Neighbour joining (NJ), Kimura 2-parameter model; substitution and partial deletion of gaps were performed with MEGA v. 5.0 (Saitou & Nei 1987, Tamura et al. 2011). Bootstrap values below 50 % were not considered. For construction of a phylogenetic tree additional sequences were obtained from GenBank based on sequences cited by Zhang et al. (2012) and Boonmee et al. (2014).
RESULTS
Isolates
Soil sample CBEC 001, which was collected from Bohani village, Madhya Pradesh, and subjected to horse hair baiting, yielded an unusual fungus. The fungus formed dark brown to black, fusoid conidia attached to beaded conidiophores with prominently swollen conidiogenous cells. The fungus was first observed after 2 mo of incubation, suggesting slow germination and growth. On SDA, the fungus was also slow growing (< 3 cm diam after 45 d).
DNA isolation, phylogeny and analysis
The partial LSU and ITS regions were sequenced for CBEC 001, and sequences were deposited in GenBank with accession no. KP765516 (ITS) and KR350633 (LSU). The ITS sequencing resulted in a 500 bp long sequence. The NCBI BLASTn result of the ITS sequence of CBEC 001 with sequences of ex-type strains of various species present in GenBank revealed 88 % similarity with Latorua caligans CBS 576.65 (KR873232; Query coverage 91 %), 86 % similarity with Polyschema sclerotigenum UTHSC-DI14-305 (KP769975; Query coverage 90 %), and 91 % similarity with Murilentithecium clematidis MFLUCC 14-0562 (KM408757; Query coverage 62 %).
The NCBI BLASTn search with the LSU sequence of CBEC 001 with sequences of ex-type strains of various species revealed 97 % similarity with Latorua grootfonteinensis CBS 369.72 (KR873267; Query coverage 99 %), 96 % similarity with Latorua caligans CBS 576.65 (KR873266; Query coverage 100 %), 96 % similarity with Polyschema sclerotigenum UTHSC-DI14-305 (KP769976; Query coverage 100 %), and 95 % similarity with Polyschema terricola CBS 301.65 (EF204504; Query coverage 100 %). The BLASTn result in the UNITE database also revealed hits with many uncultured sequences but with lesser identity. All these genera are also quite different morphologically and do not support inclusion of CBEC 001 in any of them.
TAXONOMY
Matsushimamyces Rahul Sharma, Rohit Sharma & Crous, gen. nov.
MycoBank MB810895
Etymology: Matsushimamyces; named in honour of the Japanese mycologist Takashi Matsushima (Kobe, Japan) whose fungal monographs remain a source of inspiration.
Diagnosis: Conidiophores solitary, erect, rarely branched, straight or flexuous, brown apically, smooth, beaded near conidium attachment points, constricted at the septa. Conidiogenous cells pale brown, coarsely verruculose, monoblastic, terminal. Conidia solitary, straight or curved, fusoid to broadly ellipsoid, multi-septate, apical and basal cell thin-walled, subhyaline to brown, median cells dark brown to black, thick-walled, roughly to coarsely verruculose.
Type species: Matsushimamyces bohaniensis Rahul Sharma et al. 2015.
Description: Colonies effuse, black, shortly hairy, slow growing, reverse black. Mycelium partly superficial partly immersed in the artificial substratum, composed of a network of branched, septate, pale to medium brown hyphae, smooth to sparsely verruculose, thick-walled, stroma at times present. Conidiophores macronematous, mononematous, at times aggregated, erect, unbranched (or rarely branched), straight or flexuous, hyaline to brown apically, smooth, beaded near conidium attachment, constricted at septa, intermediate cells dark brown or blackish brown, cylindrical, doliiform or lageniform. Conidiogenous cells pale brown, coarsely verruculose, monoblastic, integrated determinate, terminal. Conidia solitary, straight or curved, fusoid to broadly ellipsoid, multi-septate, apical and basal cell thin-walled, subhyaline to brown, median cells dark brown to black, thick-walled, roughly to coarsely verruculose, basal cell conic or round, truncate or apiculate.
Notes: Matsushimamyces is unique based on the following combination of characters: the solitary, pigmented conidiophores that are beaded near the terminal monoblastic conidiogenous cell, and solitary, straight or curved, fusoid to broadly ellipsoid, multi-septate conidia. The conidia are also subhyaline to brown, with dark brown to black, thick-walled, roughly to coarsely verruculose median cells, and apical and basal cells that are thin-walled, and pale brown.
Matsushimamyces bohaniensis Rahul Sharma, Rohit Sharma & Crous, sp. nov.
MycoBank MB810896
Fig. 1.
Matushimamyces bohaniensis (CBEC001 – holotype) A. Soil collection site in village Bohani. B. Close-up of colony front on SDA after 55 d showing irregular margin and carbon black colour. C. Relative size of colony on a 90 mm plate showing partly submerged and partly emerged nature. D. Reverse of colony after 55 d. E. Diagrammatic representation of the developing conidia on surface of primary culture (actual slide preparation seen in Fig 2C, X100). F. Diagrammatic representation of developing conidia on horse hair (CBEC001 – holotype). Bars: E–F = 18 μm.
Fig. 2.
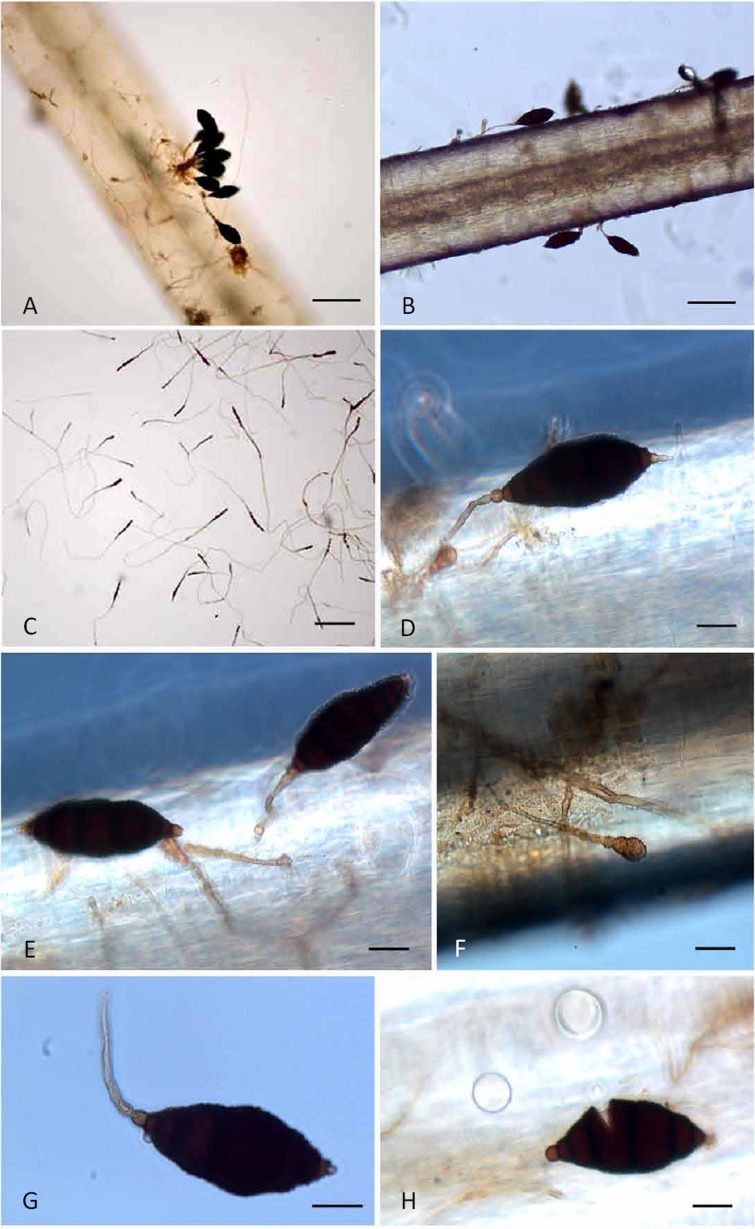
Matsushimamyces bohaniensis (CBEC 001– holotype) A. Conidia on aggregated conidiophores. B. Fungus growing and sporulating on white horse hair. C. Numerous developing conidia on SDA surface. D. Conidium showing light coloured apical and basal cell including swollen spore mother cell/conidiogenous cell. E. Detached and attached conidium from conidiophore. F. Developing conidia at a very early stage of development. G. Single conidium with attached conidiophore indicating that it is not rhexolytic dehiscence, also visible in F where conidiophore is still intact. H. Broken spore (asexual) due to external pressure exerted on cover slip showing characteristic fragmentation reminiscent of a phragmospore. Bars: A–B = 100 μm; C = 150 μm; D–H = 20 μm.
Etymology: The species epithet bohaniensis refers to the village “Bohani” (10 km from Tehsil Gadarwara, Madhya Pradesh, India) where the soil which yielded this new fungus was collected.
Diagnosis: Conidiophores macronematous, mononematous, solitary to aggregated, beaded near apex, constricted at the septa. Conidiogenous cells pale brown, verruculose, monoblastic, integrated determinate, terminal. Conidia solitary, straight or curved, fusoid to broadly ellipsoid, 6–7-septate, 50−104 × 26−33 μm, apical and basal cell thin-walled, subhyaline to brown.
Type: India: Madhya Pradesh: Bohani (22.9047°N, 79.4587°E) near Gadarwara, Narsinghpur, black conidia growing on white horse hair (keratin bait) in soil CBECS 001 collected from village garbage frequented by pigs, 16 Apr. 2014, Rahul & Rohit Sharma (CBECH 001 – holotype; CBECC 001 = MTCC12245 − ex-type cultures).
Description: Colonies effuse, black, hairy, slow growing, reaching 2 cm diam on SDA after 45 d, reverse black, with red exudate in SDA (Hi-Media ingredients), but lacking any exudate in readymade SDA or PDA (Merck). Mycelium partly superficial partly immersed in artificial substratum, composed of a network of branched, septate, pale to medium brown hyphae, smooth to sparsely verruculose, hyphae 3 μm thick, stroma at times present. Conidiophores macronematous, mononematous, solitary to aggregated, rarely branched, straight or flexuous, hyaline to brown apically, smooth, beaded near conidium attachment, constricted at septa, intermediate cells dark brown or blackish brown, cylindrical, doliiform or laginiform. Conidiogenous cells pale brown, verruculose, monoblastic, integrated determinate, terminal. Conidia solitary, straight or curved, fusoid to broadly ellipsoid, 6–7-septate, 50−104 × 26−33 μm, apical and basal cell thin-walled, subhyaline to brown, median cells dark brown to black, thick-walled, roughly to coarsely verruculose, basal cell conic or round, truncate or apiculate, apical cell round or nipple-shaped. On PDA after 40 d the conidia have big protuberances (warts), appearing abnormal with various irregular shapes.
Sexual morph: Not known.
Substratum: Baited with horse hair from soil.
Distribution: Known only from type locality, Bohani, Madhya Pradesh, India.
Matsushimamyces venustum (Castañeda et al.) Rahul Sharma, Rohit Sharma & Crous, comb. nov.
MycoBank MB814040
Basionym: Polyschema venustum R.F. Castañeda et al., Mycotaxon 57: 452 (1996); as “venusta”.
Description and illustrations: Castañeda et al. (1996).
Substratum: On decaying leaves of unidentified tree.
Distribution: Known only from type locality Jaruco, Habana province, Cuba.
Notes: Morphologically Polyschema venustum (MUCL 39001 = CBS 140212 – ex-type cultures) resembles Matsushimamyces bohaniensis in having fusiform conidia which are quite distinct from the conidial shapes of other species of Polyschema s. str. It differs from M. bohaniensis in faster growing colonies (10–15 mm in 7 days); non-beaded conidiophores and smaller conidia (30–60 × 13–20) with more septa (5–10). The new combination Matsushimamyces venustum is therefore made for P. venustum based on its conidial morphology and DNA phylogeny.
DISCUSSION
Matsushimamyces bohaniensis somewhat resembles species of the genus Manoharachariella (based on M. lignicola). Manoharachariella is distinct, however, in that it has dictyospores that frequently have a basal appendage, lacks a beaded conidiophore, and does not have a swollen apical cell. Another morphologically similar genus is Pithomyces. However, Pithomyces has ellipsoidal dictyo- and phragmospores, as well as solitary, brown, rhexolytic, sympodial denticles. Matsushimamyces differs from Bahugada in that the latter has dictyospores formed on micronematous conidiophores, with a prominantly pale brown basal cell. Hadrosporium forms a stroma with aggregated conidiophores, and has conidia that are uniformly brown except for their end cells. Another genus that needed to be considered was Septosporiopsis, as species in that genus also have dictyo/phragmospores, but the conidia in that case also have a equatorial appendage, and the conidiogenous cells proliferate percurrently. The conidia of Matsushimamyces superficially also resemble the dictyochlamydospores of Chlamydotubeufia huaikangplaensis (Tubeufiaceae; Bonmee et al. 2011), but differ in that the conidia lack vertical septa, and the new genus clusters in Latoruaceae not Tubeufiaceae. Matsushimamyces bohaniensis closely resembles Polyschema venustum, but both species are morphologically and phylogenetically quite distinct from the type species of Polyschema, P. terricola (Castañeda-Ruiz et al. 2000).
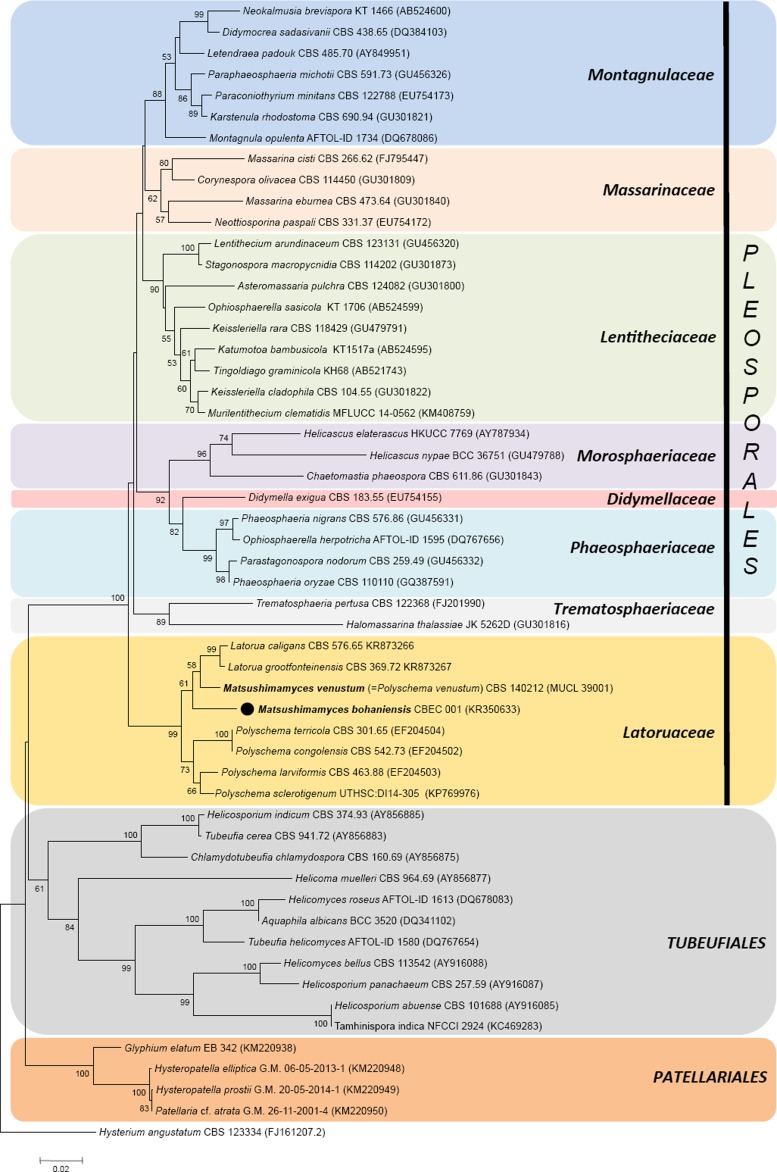
In spite of various ongoing studies, sequence databases still poorly reflect the diversity that exists in this group of fungi (Zhang et al. 2012, Boonmee et al. 2014). A neighbour-joining phylogenetic tree constructed (Fig. 3) using the LSU sequences of Matsushimamyces revealed that the genus belonged to a monophyletic lineage with Latorua and Polyschema species in Latoruaceae (Pleosporales). Matsushimamyces is separate from other families of suborder Massarineae, and forms a distinct clade of Latoruaceae (with 99 % bootstrap support) with six other members, four species of Polyschema forming a subclade and two species of Latorua (Fig. 3). The same topology was also achieved by running maximum likelihood and parsimony analyses (trees not included). In these analyses, Matsushimamyces was most closely related to Latorua (Crous et al. 2015) and Polyschema, whose species are distinguished on the basis of their conidial morphology (Castañeda-Ruiz et al. 2000). The genus Matsushimamyces differs from other genera in this complex in conidial morphology, with one apical hyaline and basal cell, hyaline to brown, beaded conidiophores, monoblastic conidiogenous cells, and typically opaque, fusoid, verruculose conidia.
Fig. 3.
A neighbour-joining phylogenetic tree constructed using 28S rDNA sequences of members of different families of Massarineae (order Pleosporales) along with Tubeufiales and Patellariales showing phylogenetic positioning of the new genus. Hysterium angustatum was used as outgroup. The analysis involved 54 nucleotide sequences and there were 575 positions in the final datasets.
ACKNOWLEDGEMENTS
We thank Narendra D Sharma for valuable discussions and constructive criticisms that helped improve the manuscript. We also thank Johannes Z Groenewald for providing sequences of Polyschema venustum (MUCL 39001), and Cony Decock (MUCL) for making this strain available for comparison. In addition, we thank Madhu Swamy (Veterinary University, Jabalpur) for allowing us to use her Nikon microscope. Financial support for this study was provided by Centre for Biodiversity Exploration and Conservation (CBEC), Jabalpur (society registration certificate no. 04/1401/16738/14) and the Department of Biotechnology (DBT), Government of India, for funding the Microbial Culture Collection (MCC), Pune (grant letter no. BT/PR10054/NDB/52/94/2007).
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Boonmee S, Rossman AY, Liu WJ, Dai DQ, Bhat DJ, et al (2014) Tubeufiales, ord. nov., interesting sexual and asexual generic names. Fungal Diversity 68: 239–298. [Google Scholar]
- Castañeda-Ruiz RF, Decock C, Saikawa M, Gene J, Guarro J. (2000) Polyschema obclaviformis sp. nov., and some new records of hyphomycetes from Cuba. Cryptogamie Mycologie 21: 215–220. [Google Scholar]
- Crous PW, Carris LM, Giraldo A, Groenewald JZ, Hawksworth DL, et al (2015) The Genera of Fungi - fixing the application of the type species of generic names – G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6: 163–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL. (1998) Getting to grips with fungal biodiversity in the tropics. In: Frontiers in Biology: the challenges of biodiversity, biotechnology and sustainable agriculture (Chou C-H, Shao K-T, eds): 57–63. Taipei: Academia Sinica. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. (2000) Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: a laboratory manual. 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sharma R, Rajak RC. (2003) Keratinophilic fungi: nature’s keratin cycling machines! Their isolation, identification and ecological role. Resonance 8(9): 28–40. [Google Scholar]
- Sharma R, Gräser Y, Singh SK. (2013) Auxarthronopsis, a new genus of Onygenales isolated from the vicinity of Bandhavgarh National park, India. IMA Fungus 4: 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Kulkarni G, Sonawane MS, Shouche YS. (2014) A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience 55: 118–123. [Google Scholar]
- Sharma R, Singh SK. (2013) A new species of Gymnoascus with verruculose ascospores. IMA Fungus 4: 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA 5.0: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Cigelnik E, O’Donnel K. (1999) Phylogeny and PCR identification of clinically important zygomycetes based on nuclear ribosomal-DNA sequence data. Journal of Clinical Microbiology 37: 3957–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crous PW, Schoch CL, Hyde KD. (2012) Pleosporales. Fungal Diversity 53: 1–221. [DOI] [PMC free article] [PubMed] [Google Scholar]