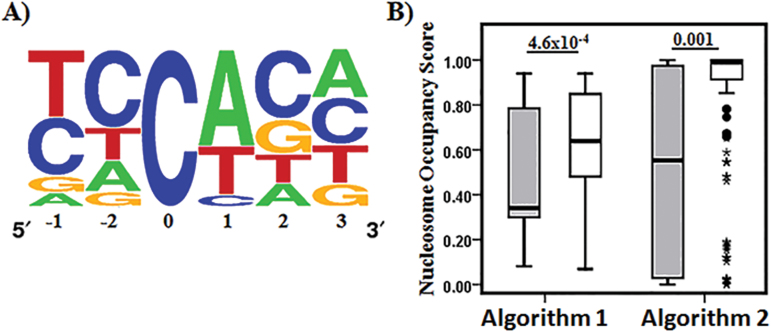
Fig. 3.
Comparison of modified and unmodified CpH sites. (A) CpA is overrepresented at modified CpH sites. Sequence logo43 generated from multiple sequence alignment of modified CpH sites (ie, CpH sites with ≥2% modification density in at least 10 individuals) with modified cytosine at position 0. (B) Nucleosome occupancy score determined for modified (grey bars) and unmodified (white bars) CpH sites by nucleosome occupancy prediction algorithm 135 and 2.36 Significantly lower score for modified CpH sites indicate they are present in nucleosome void regions of HCG9. P values above boxplots were obtained from Mann-Whitney U test.