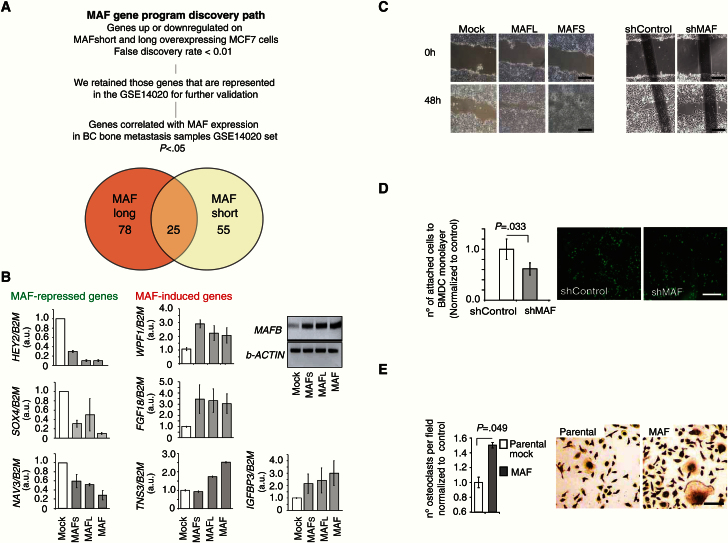
Figure 4.
MAF regulation of expression of genes associated with bone metastasis in breast cancer (BC). A) Discovery path for the identification of genes whose expression is up- or down-regulated in MAF short- and/or long-expressing cells compared with parental Mock-MCF7 cells. Pearson correlation and fold change bigger than two and Bayesian false discovery rate below 5%. B) Expression levels of genes identified in (A) parental control, MAF-short and MAF-long isoform-overexpressing cells measured by quantitative real-time polymerase chain reaction using indicated TaqMan or SybrGreen probes. B2M or b-ACTIN were used as normalization controls. Data is mean of three experiments with SD. C) Representative images from three independent experiments show wound-closing assay at initial and end timepoints. The extent of wound closure was assessed after 48 hours. Scale bar = 200 μm. D) Representative image of attached green-labeled tumor cells to monolayer of BMSC. Data from three independent experiments is presented as average with SD. P value scored by two-sided Wilcoxon signed-rank test. Scale bar = 500 μm. E) Quantification of the number of TRAP+ differentiated multinucleated (>3 nuclei) osteoclasts per field, normalized to control; representative images are shown. Scale bar = 100 μm. Conditioned media from the indicated cellular populations were used. Data are means from three independent experiments with SD. P value scored by two-sided Wilcoxon signed-rank test.