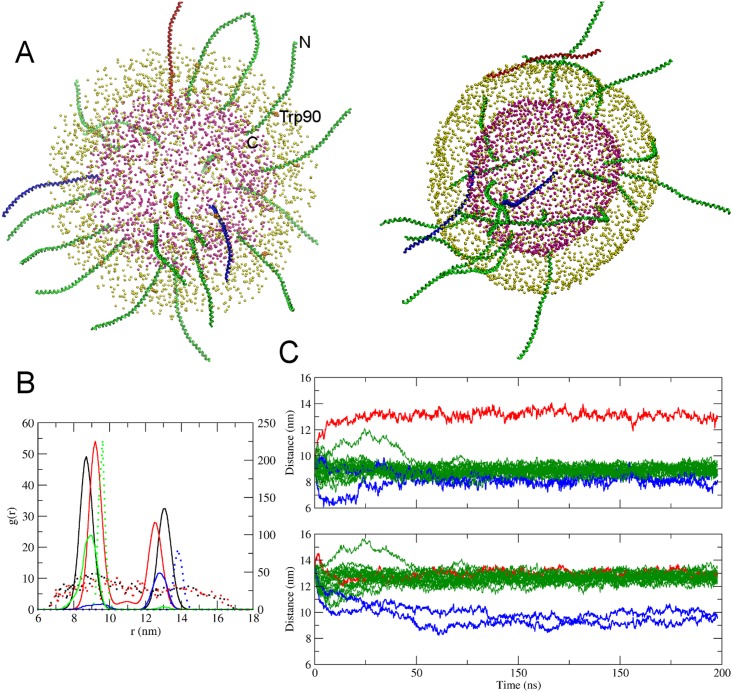
Fig 9. Self-assembly of the vesicle-protein system.
(A) Snapshot of vesicle at the start (left) and at the end (right) of self-assembly done along with twenty copies of syb2 molecules. To aid visualization, only the phosphate headgroups of the lipids and the backbone beads of the protein are shown. The phosphate headgroups of the outer leaflet are shown in yellow and the inner leaflet in magenta. The backbone beads of Syb2 copies that positioned themselves correctly are shown in green; three syb2 molecules that failed to position themselves correctly are colored red and blue. To illustrate the initial positioning of syb2, the left panel also shows theTrp90 in vdw representation and colored orange. The N- and C-term is also labeled for one of the syb2 molecule. (B) Comparison of the RDF g(r) of the lipid phosphate groups (black), cholesterol (red), Thr116 (green) and Trp90 (blue) at the start of the simulation (dotted) and during the last 50 ns of the self-assembly (solid). Note the small RDF peak of Thr116 (green solid) near the outer leaflet phosphate headgroup (red molecule in A) and the appearance of a Trp90 peak (blue solid) near inner leaflet phosphates (blue molecules in A), originating from misoriented syb2 molecules. (C) Evolution of the distance of backbone beads of Thr116 (top) and Trp90 (bottom) residues of all twenty protein molecules from the center of mass of the forming vesicle. The traces are colored following the coloring scheme of the molecules as shown in A.