Abstract
Large-scale sequencing efforts are uncovering the complexity of cancer genomes, which are comprised of causal “driver” mutations that promote tumor progression along with many more pathologically-neutral “passenger” events. The majority of mutations, both in known cancer drivers and uncharacterized genes, are generally of low occurrence, highlighting the need to functionally annotate the long tail of infrequent mutations present in heterogeneous cancers. Here we describe a mutation assessment pipeline enabled by high-throughput engineering of molecularly-barcoded gene variant expression clones identified by tumor sequencing. We first used this platform to functionally assess tail mutations observed in PIK3CA, which encodes the catalytic subunit alpha of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) frequently mutated in cancer. Orthogonal screening for PIK3CA variant activity using in vitro and in vivo cell growth and transformation assays differentiated driver from passenger mutations, revealing that PIK3CA variant activity correlates imperfectly with its mutation frequency across breast cancer populations. While PIK3CA mutations with frequencies above 5% were significantly more oncogenic than wild-type in all assays, mutations occurring at 0.07 – 5.0% included those with and without oncogenic activities that ranged from weak to strong in at least one assay. Proteomic profiling coupled with therapeutic sensitivity assays on PIK3CA variant-expressing cell models revealed variant-specific activation of PI3K signaling as well as other pathways that include the MEK1/2 module of Mitogen-Activated Protein (MAP) Kinase pathway. Our data indicate that cancer treatments will need to increasingly consider the functional relevance of specific mutations in driver genes rather than considering all mutations in drivers as equivalent.
Keywords: Cancer genomics, functional screens, mutagenesis, molecular barcoding, breast cancer, PIK3CA
INTRODUCTION
Next generation sequencing (NGS) provides a powerful tool for elucidating the genetic underpinnings of tumor initiation and progression. A central goal of large-scale projects such as The Cancer Genome Atlas (TCGA) is to catalog the genomic landscape of diverse cancers while illuminating new diagnostic biomarkers and therapeutic targets. Such efforts have confirmed recurrent signature mutations in cancer driver genes that include KRAS, NRAS, BRAF, PIK3CA, EGFR, TP53, and PTEN among others. Knowledge on the functional impact of these events, efforts toward developing therapies targeting their activity and plummeting sequencing costs have prompted healthcare providers to incorporate NGS into the clinic to facilitate diagnostic and therapeutic decisions. As we continue in the era of precision medicine, physicians will have increased access to patient tumor genomes, ultimately providing a means to subtype disease, provide rational use of molecular-guided therapies and monitor disease progression. Mainstream cancer protocols assign patients to a particular treatment regimen based predominately on their diagnosed cancer, yet modernized approaches call for choosing therapeutics based on the underlying mutations in each individual patient’s tumor.
While much is known about the function and clinical impact of recurrent “hotspot” mutations in genes including those listed above, less is known about which and how the more abundant, low-frequency mutations contribute to tumor progression. This is true for not only mutations in under-described or novel genes, but also rare “tail mutations” in well-studied cancer genes often mutated at hotspot positions. For example, the p110α catalytic subunit of the phosphatidylinositol 3-kinase (PI3K) encoded by PIK3CA represents the most commonly mutationally-activated gene in the cancer genome (1). In breast cancer, the most frequent PIK3CA mutations lead to protein residue changes at positions E542, E545 and H1047, comprising approximately 78% of all PIK3CA mutations observed in breast tumors (2,3). The high frequency of these hotspot mutations has prompted much investigation into their functional impact, thus it is well-established that they lead to gain-of-function PI3K activity, downstream signaling through the AKT/mTOR pathway and robust in vitro and in vivo transformation phenotypes (4–7). As a result, much focus has been placed on developing PI3K pathway inhibitors that are at varying stages of clinical testing in several cancer types.
In addition to these PIK3CA hotspot mutations, the sequencing of greater numbers of tumors is revealing numerous tail mutations existing outside the hotspot locations. While some efforts have been made to functionally characterize PIK3CA tail mutations [e.g., (8–11)], their growing number among tumor NGS datasets requires flexible approaches to characterize the functional impact of large numbers of mutant variants. This is true not only for PIK3CA, but also for other cancer genes with increasing numbers of identified mutations afforded by greater NGS sequencing depths. Recent studies demonstrate the need for comprehensive functional testing of cancer gene variants. For example, mutations in isocitrate dehydrogenase (IDH1 and IDH2), which occur in low-grade gliomas and secondary high-grade gliomas, inhibit the enzyme’s ability to catalyze conversion of isocitrate to alpha-ketoglutarate while conferring a neomorphic activity for reduction of α-ketoglutarate to d-2-hydroxyglutaric acid (2-HG) (12). 2-HG is proposed to function as an oncogenic metabolite by inducing hypermethylation of histones and DNA (13), thus much work is focused on developing effective inhibitors of mutant IDH1/2. Similar to studies on IDH1/2, a recent examination of mutant isoforms of PIK3R1, which encodes the p85α regulatory subunit of PI3K, revealed novel activity for cancer-associated variants (14). Unlike other mutations in PIK3R1 that function to activate PI3K activity, truncating variants at positions R34 and L370 confer unexpected neomorphic activation of JNK pathway and response to JNK inhibitors resulting from the mutant proteins’ mislocalization to the nucleus (14).
The development of driver prediction algorithms provides one means to detect oncogenic mutations in silico, though the success of these tools has been limited without validation and iterative improvement using functional screening approaches. However, experimental annotation of rare mutations is difficult given their large number, examination of which requires robust pipelines permitting scalable gene variant construction and cancer context-specific screening for activity. Here we describe a mutation assessment pipeline first used to functionally annotate tail mutations within PIK3CA discovered in breast cancer. We employ High-Throughput Mutagenesis and Molecular Barcoding (HiTMMoB) technology to construct an allelic series of PIK3CA variants, each uniquely barcoded with a molecular DNA tag allowing individual and pooled examination in vitro and in vivo to identify mutations exhibiting gain-of-function cancer activity. Expanding our analysis of the PIK3CA allelic series to functional proteomics through use of Reverse Phase Protein Arrays provided a comprehensive view of variant-induced changes to key cancer signaling pathways, dependence on which was validated by cell-based inhibitor assays. Our data indicate that many but not all PIK3CA variants similarly activate the PI3K signaling pathway, and furthermore that subsets of PIK3CA variants also trigger activation of the MAPK pathway as well as evoke cell invasion activity. When applied more broadly to other oncogenes and tumor suppressor genes, functional assessment of low-frequency mutations by use of HiTMMoB and comprehensive phenotyping will allow scalable characterization of gene variant activity with potential clinical outcomes.
MATERIALS AND METHODS
Bioinformatic genetic analysis
The Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic, queried 12/18/2014). We applied the following algorithms: SIFT (15), PolyPhen (16), MutationAssessor (17), VEST (18), CHASM (19), MutationTaster (20), Condel (21), and CanDrA (22). Exome Aggregation Consortium database (ExAC; Exome Aggregation Consortium Cohort; http://exac.broadinstitute.org, 05/05/2015) was queried to determine the germline population allele frequency of PIK3CA variants in global human populations. All variants are reported relative to PIK3CA NM_006218.2 and NP_006209.2.
Cell culture
MCF10A cells were obtained from the American Type Tissue Collection (ATCC) and HMLER cells were provided by S. Mani (MD Anderson Cancer Institute). MCF10A and HMLER cells were cultured in MEGM medium without antibiotics and serum. MCF10A cells were supplemented with 100ng/ml cholera toxin (Sigma). Ba/F3 cells were grown in RPMI medium supplemented with 5% Fetal Bovine Serum. Parental Ba/F3 cells were grown in the presence of IL3 at 5 ng/ml. Stable Ba/F3 cells expressing PIK3CA variants were always grown without IL3. MCF10A cells and HMLER cells were fingerprinted prior to use on 10/14/2014 and 4/21/2015, respectively, by the MD Anderson Cancer Center Characterized Cell Line Core using STR testing platform. Ba/F3 is a mouse-originated cell line, thus STR testing could not be performed.
Immunoblot analysis
Cells were lysed using RIPA buffer (Sigma) containing Protease Inhibitor Cocktail (Sigma) with phosphatase inhibitors (Calbiochem) for separation on NuPAGE-Bis-Tris gels (Life Technologies) and blotting onto PVDF membrane. The following antibodies were used for immunoblotting: p110alpha (Cell Signaling), phospho-MEK1/2 (Cell Signaling), phospho-p38 (Cell Signaling), phospho-S6K (Cell Signaling), phospho-S6 (Cell Signaling), phospho-AKT (Cell Signaling), phospho-PRAS40 (Cell Signaling), Vinculin (Santa Cruz).
RPPA analysis
RPPA method has been described previously (23). Cells were plated as singlets and treated growth medium without insulin and EGF for 60 hours and then lysed with RPPA lysis buffer. Then, lysate concentrations were adjusted and the lysates were mixed with SDS sample buffer.
Cloning
Site-directed mutagenesis was based on modified application of megaprimer PCR (24). For barcoding, a set of barcode entry clones was constructed by annealing in vitro complimentary DNA oligonucleotides containing 24 nt barcodes flanked by T3/T7 sequences and attB2r/B4 sites. Annealed oligonucleotides were subsequently recombined into modified pDONR223 by BP recombination, leading to entry clones whose barcodes were flanked with attR2/L4 for multi-fragment recombineering (with ORF entry clone or mutagenesis PCR) into pLenti6 V5/Dest BC that contains attR1 and attR4 recombination sites. STBL3 (Life Technologies) bacteria were heat-shock-transformed with the BP/LR reaction mixtures for vector propagation.
Stable cell lines
Lentivirus was produced using standard virus packaging vectors and virus protocols. For MCF10A and HMLER transduction, cells were infected twice with 10–12 hour periods in the presence of 8 μg/ml polybrene before blasticidin selection (10 μg/ml). Ba/F3 cells were spin-infected for 2 hours at 300g and then incubated at 37ΰC for 10 hours before refreshing the regular medium with 1:10000 diluted IL3 concentrations.
Survival assays
MCF10A cells were trypsinized and 300 or 1200 cells were seeded in 96-well opaque plates for No Insulin and No Insulin-No EGF assays, respectively, in quadruplicates with corresponding medium conditions. Cell viability was measured after 5 days for No Insulin assay and 10 days for No Insulin-No EGF assay with Cell Titer Glo (Promega) following manufacturer’s instructions. Ba/F3 IL-less survival assays were performed by removing medium after viral infection and culturing cells in low IL3 medium (1:10000 dilution) in duplicates. Cell viability was measured after 7 days.
Drug sensitivity assays
Drug experiments for MCF10A cells were done in No Insulin-No EGF condition with 1000 cells in 96-well white plates (PerkinElmer). Drugs were added at the specified concentrations the next day after seeding the cells. For Ba/F3 cells, drugs were tested in the presence or absence of IL3 and drugs were added the same day. Cell viability was measured 3 days after adding drugs with Cell Titer Glo for both cell types. All inhibitors were purchased from Selleck.
In vitro cell invasion and anchorage independent growth assays
Cell invasion assays and anchorage-independent growth assays were performed as described previously (25). Colonies were stained with 0.05% iodonitrotetrazolium chloride (Sigma) and counted after scanning with a flatbed scanner. All data were assessed by two-tailed t-test calculation using Prism 6 (Graphpad).
Animal studies
All studies using mice were performed in accordance with our IACUC-approved animal protocol at Baylor College of Medicine. 8X105 HMLER cells transduced with GFP, PIK3CAWT, PIK3CAE545K or PIK3CAH1047R were re-suspended in a 1:1 solution of Hank’s balanced salts and Matrigel (BD Bioscience) and then subcutaneously injected into female nude animals (Harlan, Indianapolis, IN) as described previously (25). For PIK3CA variant competition assays,1X105 cells from each of the HMLER cell lines expressing GFP, PIK3CAWT, eight PIK3CA constructs with silent mutations and twenty rare PIK3CA mutants were pooled and injected subcutaneously as above. Tumor volumes were calculated by following formula: Length × Width2/2
Barcode sequencing
Genomic DNA was extracted from pool of HMLER cells that were injected (Input) and 3 individual tumor cores (output) for triplicate and duplicate, respectively, PCR reactions to amplify the barcode pools present within each sample using Platinum Super Mix (Life Technologies) and flanking primers common to each barcode: 5′-CAATTAACCCTCACTAAAGG and 5′-CCGCCACTGTGCTGGATA). Amplification parameters were as follows: 1 × [94°C-4′]; 35 × [94°C-1′, 60°C-1′, 68-1′]; 1 × [68°C-10′]. Each replicate PCR amplicon (211 nt) was purified for PGM library preparation where each amplicon was ligated to unique Ion Xpress Barcode Adaptors for PGM sequencing (318 V2 Chip) following the manufacture’s recommendations. Raw data were concatenated into one ‘reference’ file and indexed using BWA for alignment of barcode sequences (with parameters “-l7 -t12 -N -n3”) for counting the occurrence of each barcode. For 2D barcode representation graph, total reads of barcodes in all tumors were averaged for each barcode and their ratio to total number of barcode reads in all tumors were plotted. Barcode enrichment for individual tumors was assessed by quantitating the number of occurrences for each barcode sequence as a ratio to total number of barcode reads in each sample. Standard deviations were calculated for triplicate and duplicate reactions for input and output samples, respectively, and plotted as error bars on the barcode enrichment graphs.
RESULTS
Selection and construction of PIK3CA tail mutations
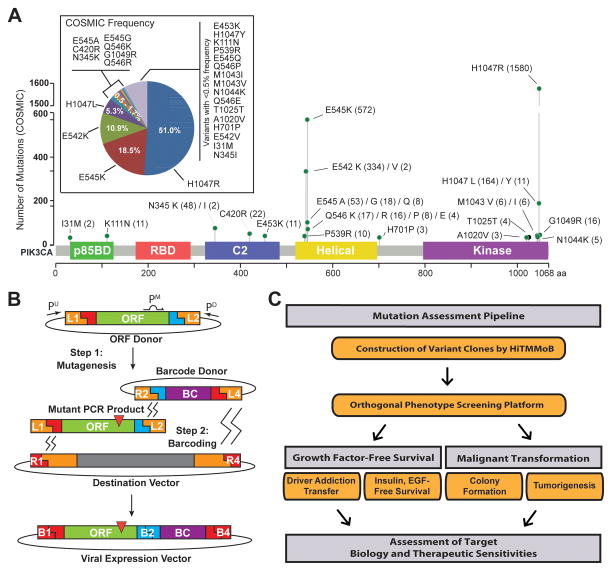
Interrogation of the Catalogue of Somatic Mutations in Cancer (COSMIC) revealed 3,096 somatic DNA aberrations in PIK3CA residing in breast cancer tumors and tumor-derived cell lines (Supplementary Table S1). The majority of these somatic events were nucleotide missense substitutions (3,063; 98.9%), and the remaining aberrations consisted of small insertions/deletions (0.68%), silent (0.29%) and nonsense substitutions (0.032%). Given the high recurrence of PIK3CA missense mutations corresponding to alterations at amino acid residues E545 (21.5%) and H1047 (57.3%), the 3,063 missense mutations encompassed only 106 variants that resulted in a single amino acid change at 75 of the 1068 residues of the encoded PIK3CA protein, p110alpha. In order to assess the functional impact of rare PIK3CA mutations, we selected 25 representative variants whose breast cancer COSMIC frequencies ranged 0.07% (present in two specimens) to 10.9% (present in 334 specimens), as well as the E545K and H1047R hotspot mutations that represent 18.5% and 51.0% of PIK3CA mutations, respectively (Fig. 1A). The 27 mutations chosen for functional annotation targeted a total of 16 amino acid residues positioned within or proximal to the p85 binding, C2, Helical and Kinase domains of p110alpha (Fig. 1A). It is noteworthy that one selected somatic mutation, T1025T, is a synonymous change residing distal to the kinase domain with breast cancer COSMIC frequency 0.1%.
Figure 1. PIK3CA variant selection and mutation assessment platform.
(A) COSMIC PIK3CA variant frequencies and distribution across domain-annotated p110α protein structure for mutations chosen for modeling and functional annotation. Number in parentheses indicates numbers of samples found mutant in COSMIC. (B) Illustration of HiTMMoB-mediated mutagenesis and molecular barcoding using megaprimer PCR and multi-fragment recombination. Recombination of mutant PCR product and barcode into destination vector denoted by zig-zag lines. ORF = open reading frame clone; BC = Barcode; Pu/Pm/PD, PCR primers; L/R/B, att recombination sequences; red triangle = mutation. See Methods for details. (C) Illustration of mutation assessment pipeline workflow.
Our selection of PIK3CA mutations was based on an unbiased analysis of breast cancer COSMIC data. In order to gain initial insight into the potential impact of these mutations, we evaluated the 26 missense mutations by 9 computational algorithms designed to predict the functional impact of mutations on protein function (Supplementary Table S2). We observed a general disagreement among the prediction algorithms, though each PIK3CA missense variant was predicted to have functional consequence by at least 4 of the 9 algorithms used. For example, there was a general consensus among algorithms that hotspot mutation E545K would result in a deleterious, possibly damaging alteration considered to be a disease-causing driver event. In contrast, hotspot mutation H1047R was considered a deleterious, disease-causing driver event by some algorithms yet neutral or benign by others. Likewise, I31M, which was identified in only two (0.07%) COSMIC samples with mutant PIK3CA, was determined to be a neutral, benign event by some algorithms yet others identified I31M as a driver mutation with a high likelihood of being a disease-causing event. The inconsistencies observed across these prediction algorithms further illustrate the need for functional examination of individual mutations within bona fide cancer drivers.
We next employed HiTMMoB to engineer mutations into the PIK3CA open reading frame (ORF) using megaprimer-based mutagenesis (26), followed by recombination-based cloning of mutant PCR products into Gateway (Life Technologies)-compatible destination vectors that enable ORF virus production for expression in mammalian cells (see Methods). Our multi-fragment recombineering cloning strategy (Fig. 1B) also permits rapid simultaneous tagging of individual ORF clones with unique 24-nucleotide DNA “barcodes”. The addition of barcodes (BCs) permits ORF pooling and tracking, as each BC serves as surrogate identifier for its associated ORF. We applied these cloning strategies to engineer wild-type, the selected 27 mutations and an additional 9 silent mutation controls (e.g., E545E and H1047H; Supplementary Table S3). HiTMMoB achieved 62.4% mutation efficiency per reaction (Supplementary Table S3), permitting clonal isolation and full sequence confirmation of individual mutant ORFs after picking an average of 3 colonies per each mutagenesis reaction. In total, we successfully constructed all 37 barcoded PIK3CA ORF clones in a single pass for functional assessment using our mutation assessment pipeline (Fig. 1C).
PIK3CA tail mutations promote Ba/F3 survival
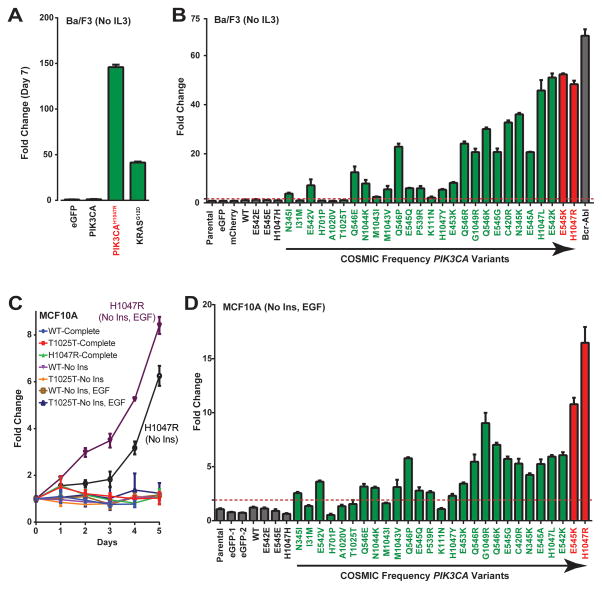
Many studies have characterized the phenotypic consequence of expressing PIK3CA hotspot variants, and some effort has been made to rare PIK3CA mutant effects on kinase activity and transformation of chicken and mouse fibroblast models among others [e.g., (8–11); Supplementary Table S4]. In addition to malignant transformation, activating mutations in oncogenes such as PIK3CA are known to promote sustained proliferation and survival in the absence of growth factors. We modeled these hallmarks of cancer by entering our PIK3CA allelic series into assays to assess growth factor-free survival. We first employed the Ba/F3 murine, IL3-dependent pro-B cell model (27) used by us (14,28) and others to screen for the ability of driver genes to induce cell survival and proliferation. Ba/F3 cells die in the absence of exogenous IL3; therefore, we quantitated cell growth as a measure of “transfer of driver addiction” from IL3 to PIK3CA variant ORFs and controls following IL3 removal from growth medium. For example, expression of hotspot mutant ORFs PIK3CAH1047R and KRASG12D promote robust Ba/F3 cell proliferation in the absence of IL3 compared to undetectable growth by cells expressing green fluorescent protein (eGFP; negative control) or wild-type PIK3CA at 7 days following removal of IL3 (Fig. 2A). Analysis of PIK3CA variants in Ba/F3 confirmed potency for PIK3CAH1047R as well as a >40-fold increase in growth-promoting activity for high-frequency (>5.0%) PIK3CA mutations E542K, E545K, H1047L and H1047R (Fig. 2B). In contrast, cells expressing wild-type PIK3CA or silent controls (E542E, E545E, H1047H) proliferated similar to parental Ba/F3 cells and those expressing eGFP and mCherry (Fig. 2B). Of the remaining 23 low-frequency variants (<5.0% of PIK3CA mutations in COSMIC breast cancer data), 19 promoted Ba/F3 survival and proliferation at a range of 2.1- to 36-fold over the mean of negative controls (parental, eGFP, mCherry, silent mutations). Nine of 12 driver events with a COSMIC frequency less than 0.5% (represented ≤ 11 of 3,096 PIK3CA mutations; E542V, Q546E, N1044K, M1043V, Q546P, E545Q, P539R, H1047Y and E453K) exhibited a significant >5-fold impact on Ba/F3 IL3-indendent survival despite their low frequency, indicating that ultra-rare PIK3CA mutations can provide driver activity. Spearman’s correlation analysis of these data revealed that the fold-change increase in Ba/F3 growth significantly correlated (r=0.83, p=3.6E-08) with increasing COSMIC frequency of PIK3CA variants; however, this correlation is imperfect as some low-frequency mutations were more active than more common events (e.g., Q546E and Q546P). This correlation was not entirely due to hot-spot mutations PIK3CAE545K and PIK3CAH1047R (69.5% combined COSMIC frequency), as the Spearman’s p-value remained significant (r=0.79, p=1.2E-06) following removal of the E545K and H1047R datasets from the Spearman’s analysis. Four PIK3CA variants (H701P, I31M, T1025T and A1020V) exhibited no activity in Ba/F3, which suggests that these events may represent true passenger mutations as assessed under the conditions and driver sensitivity of this assay.
Figure 2. PIK3CA tail mutations promote growth factor-free survival.
(A–B) Ba/F3 cell survival in the absence of IL3 for (A) the indicated controls and (B) the expanded series of negative controls (gray), rare (green) and hotspot (red) PIK3CA variants. Bcr-Abl = positive control for Ba/F3 assay. (C) Growth curve for MCF10A cells expressing wild-type (WT) or the indicated PIK3CA variants (PIK3CAH1047 and PIK3CAT1025T) in the presence or absence of growth factors insulin (Ins) and EGF. Complete = growth factor-complete medium. (D) MCF10A cell lines expressing the indicated PIK3CA variants assessed for cell growth in the absence of insulin and EGF. All data in B and D normalized to the mean of the negative controls. Dashed line = significance limit set as value equal to 3x standard deviations of negative control values. Data normalized to the average of negative controls. Error bars depict standard deviation.
As a means of providing orthogonal evidence for the notion of driver versus passenger mutation, we determined the allele frequencies of the 27 engineered and functionally vetted variants in across global human populations. The ExAC database contains exome data generated from the germlines of 60,000 adults representing Europe, Africa, Asia, and America. Notably, our analyses revealed that PIK3CA c.C3075T (p.T1025T), is a high frequency variant in the human germline ranging from as high as 13% of South Asians to a low of 0.2% of East Asians. This finding is consistent with our experimental evidence for T1025T being a passenger mutation. The only other tested variants observed in ExAC were c.A3140G (p.H1047R), c.A3140T (p.H1047L), c.G1633A (p.E545K), each of which were observed in only one chromosome out of 120,000. We found no indication that the remaining three inactive variants (H701P, I31M and A1020V) represented common germline variants, which further illustrates the need to for functional evaluation of all variants of unknown significance for a given disease-associated gene.
PIK3CA tail mutations promote growth factor-free survival of breast epithelial cells
While serving as a tractable cell model for generalized evaluation of driver activity, Ba/F3 cells may not provide the optimal cell lineage context for analysis of breast cancer-specific mutations and may only detect a subset of oncogenic events. To examine this further, we assessed PIK3CA variant activity using normal MCF10A breast epithelial cells widely used to assess the oncogenic impact of breast cancer drivers. MCF10A cell growth is dependent on insulin and epidermal growth factor [EGF; Supplementary Fig. S1A], which are both potent activators of signaling pathways activated in diverse cancer types. MCF10A cells stably expressing PIK3CAH1047R exhibited a 6.0- and 8.2-fold increase in cell growth in culture medium lacking insulin (p<0.0001) and medium lacking insulin and EGF (p<0.0001), respectively, whereas wild-type- and PIK3CAT1025T-expressing control cells did not exhibit a growth advantage under matched-normalized conditions (Fig. 2C). In contrast, no growth difference was observed between the cell lines when propagated in medium fully supplemented with insulin and EGF (Fig. 2C). Expanding this analysis to the PIK3CA allelic series confirmed potent growth-promoting activity for H1047R (16.5-fold, p<0.0001) and E545K (10.8-fold, p<0.0001) hotspot mutations in the absence of insulin and EGF relative to the mean of all negative controls (Fig. 2D). The remaining PIK3CA variants performed similar to our findings using Ba/F3, exhibiting a 2.3- to 9.0-fold range increase in MCF10A growth that correlated, albeit imperfectly, with each variant’s frequency in COSMIC (with hotspot data: r=0.75, p=3.0E-06; without hotspot data: r=0.68, p=7.3E-05) whereas cells expressing wild-type PIK3CA and silent controls proliferated similar to parental and eGFP-expressing MCF10A cells (Fig. 2D). PIK3CA variants H701P, I31M, T1025T and A1020V did not provide a cell growth advantage in MCF10A, which is consistent with results using Ba/F3, further suggesting they may represent passenger mutations. Moreover, our data indicate that PIK3CAG1049R (COSMIC frequency = 0.5% or 16 of 3,096 PIK3CA mutations) activity was greater than all other low-frequency variants, exhibiting a 9.0-fold change increase in MCF10A growth similar to the PIK3CAE545K mutation with COSMIC frequency of 18.5%. On the other hand, expression of PIK3CAM1043I and PIK3CAK111N provided no growth advantage to MCF10A cells in the absence of insulin and EGF despite exhibiting driver activity in Ba/F3 cells. Assaying MCF10A growth in the presence of EGF (No insulin) revealed growth-promoting activity for the M1043I and K111N variants but not H701P, I31M, T1025T and A1020V (Supplementary Fig. S1B), suggesting that the observed MCF10A and Ba/F3 discordance for PIK3CAM1043I and PIK3CAK111N might be due either to increased assay stringency upon combined removal of insulin and EGF conditions or specific insulin pathway dependencies for these two variants.
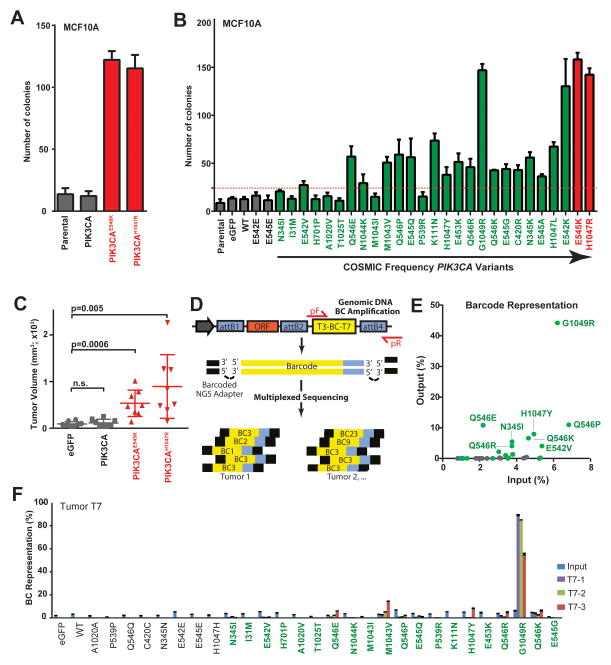
PIK3CA tail mutations drive malignant transformation
Assaying the ability of oncogene expression to promote colony formation in semi-solid culture medium is widely used to assay for anchorage-independence, a hallmark of cell transformation (29). Therefore, we next entered the PIK3CA allelic series into MCF10A soft agar colony formation assays to determine each variant’s ability to promote cell transformation in vitro. Similar to as reported by others (5), stable expression of both PIK3CAH1047R and PIK3CAE545K led to a significant increase in colony formation (9.5- and 9-fold, respectively, p<0.0001; Fig. 3A) compared to wild-type PIK3CA-expressing and parental control cells. Expanding the colony formation assay to the PIK3CA allelic series revealed potent activity for high-frequency mutations E545K and H1047R (average 11.8-fold increase over negative controls, p<0.0001) and a range of activities (2.3-fold to 11.5-fold) for PIK3CA variants (Fig. 3B; Spearman’s correlation analysis with COSMIC frequencies with hotspot data: r=0.67, p=6.3E-05; without hotspot data: r=0.59, p=8.6E-04). PIK3CA variants H701P, I31M, T1025T and A1020V did not promote colony formation, which is consistent with their inactivity in the growth factor-free survival assays using Ba/F3 and MCF10A (Fig. 2). We also did not observe colony-forming activity for PIK3CA variants N345I, M1043I and P539R, which exhibited very weak activity in the growth factor-free survival assays (Fig. 2), a discordant result potentially due to the increased biological stringency of the colony transformation assay. As noted for MCF10A growth factor-free survival (Fig. 2D), the low-frequency G1049R variant promoted colony formation similar to the hotspot mutations E545K and H1047R, further supporting the notion that this mutation is a rare but strong oncogenic PIK3CA mutation.
Figure 3. PIK3CA tail mutations promote in vitro cell transformation and oncogenic tumor growth (A–B).
MCF10A anchorage-independent colony formation assays for (A) the indicated controls and (B) the expanded series of negative controls (gray), rare (green) and hotspot (red) PIK3CA variants. Dashed line = significance limit set as value equal to 3x standard deviations of negative control values. Data normalized to the average of negative controls. (C) Endpoint volumes (day 36 post-injection) for xenograft tumors resulting from HMLER cells expressing control (eGFP and wild-type PIK3CA; gray) and hotspot variants (PIK3CAE545K and PIK3CAH1047, red). P-value calculated by t-test; n.s. = not significant. (D) Illustration of barcode design and NGS sequencing strategy. Large arrow, vector promoter; pF/pR = forward and reverse PCR amplicon primers; ORF = open reading frame clone; BC = Barcode; attB1/2/4, recombination sequences. See Methods for details. (E) Two-dimension dot plot indicating the ratio of barcode reads for each negative control (gray) and rare (green) variant barcode in injected cells (Input, X-axis) in all tumor samples (Y-axis) to total number of barcode reads. (F) Barcode enrichment analysis of representative tumor, T7. Error bars represent ratio of number of barcode reads for mutations to total number of barcode reads from 3 (T7-1, 2, 3) cores of a single experimental tumor, T7. Error bars depict standard deviation of sequencing amplicon replicates.
We next sought to evaluate the oncogenic activity of rare PIK3CA variants by performing xenograft tumorigenesis assays in mice. We chose to evaluate tumor driving activity by modeling our PIK3CA allelic series into primary human mammary epithelial cells (HMECs) having undergone sequential retroviral-mediated expression of the telomerase catalytic subunit, SV40 large T and small t antigens and H-RasV12 [HMLER cells, (30)]. HMLER cells are primed for tumorigenesis and do not form tumors unless H-RasV12 levels are substantially high, suggesting they require more cooperating events to drive tumor formation (30,31). Consistent with this notion, stable expression of hotspot variants PIK3CAH1047R and PIK3CAE545K robustly drive tumor growth by HMLER when implanted subcutaneously into athymic mice, whereas control cells stable expressing eGFP or wild-type PIK3CA fail to drive frank tumorigenesis (Fig. 3C). Based on this finding we next performed a competitive tumorigenesis assay whereby equal numbers of HMLER cell populations, each engineered to express an individual barcoded PIK3CA variant ORF (COSMIC frequency ≤ 0.5%), along with an additional 10 control cell populations (barcoded ORFs encoding eGFP, wild-type PIK3CA and 8 PIK3CA silent mutation controls) were implanted into a cohort of 15 athymic mice. We reasoned that highly oncogenic PIK3CA variants would be positively enriched in tumor tissue over weak variants or those providing no transforming potential. Based on this precept, we employed NGS (Fig. 3D) barcode enrichment analysis to quantitate the number of barcode sequencing reads in tumor (i.e., “output”; 3 tumor cores per 7 tumors and technical replicates for each sample) and injected cells (i.e., “input”; 2 input replicates), followed by comparing the number of individual barcode reads between populations as a ratio of each barcode to the total number barcode reads per sample. Analysis of all tumor cores (Supplementary Table S5) indicated high concordance with the MCF10A colony formation data (Fig. 3B), revealing PIK3CAG1049R as the most significantly-enriched, rare PIK3CA ORF across all tumors (44.2% of all tumor barcode reads; Fig. 3E). We observed less enrichment for N345l, E542V, Q546E/R/K/P and H1047Y (average 7.1%), which is consistent with their relatively weaker colony-formation activity (Fig. 3B) compared to G1049R. At the individual tumor level (representative tumor T7 cores 1–3 shown in Fig. 3F), we observed a range of heterogeneous variant complexities in the backdrop of high PIK3CAG1049R enrichment.
Characterization of PIK3CA pathway activation through functional proteomics
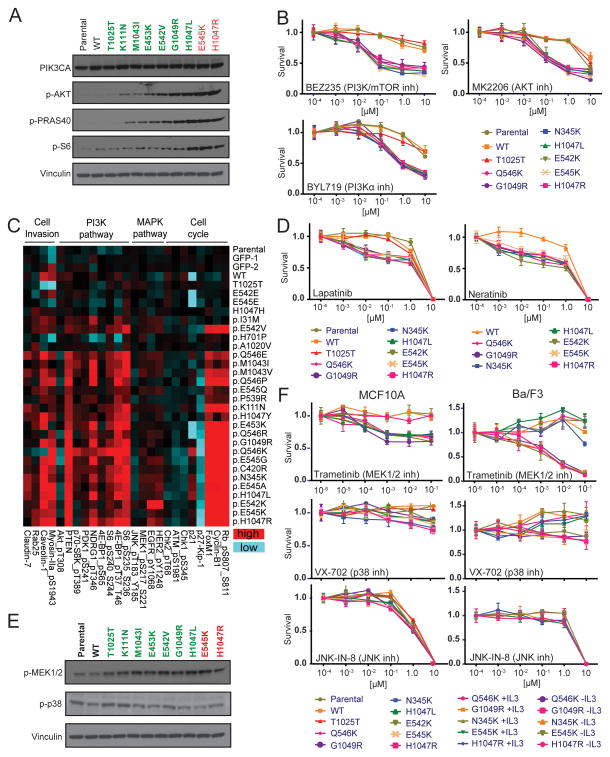
PIK3CA encodes the 110α is catalytic subunit of PI3K, and highly oncogenic mutations in PIK3CA such as H1047R and E545K result in ligand-independent activation of the PI3K/AKT/mTOR signaling pathway (32). In order to determine whether rare PIK3CA variants similarly activate PI3K signaling, we examined the phosphorylation levels of key pathway intermediates AKT, PRAS40, and S6. Immunoblot analysis of growth factor-deprived MCF10A cells expressing hotspot variants PIK3CAH1047R and PIK3CAE545K confirmed robust activation of PI3K/AKT/mTOR compared to parental MCF10A cells and those expressing wild-type PIK3CA and PIK3CAT1025T (Fig. 4A). As observed in the phenotypic profiling studies (Figs. 2–3), expression of the rare but highly active variant PIK3CAG1049R led to greater pathway activation compared to phenotypically weaker mutations that include K111N, M1043I, E453K and E542V (Fig. 4A). Consistent with these observations, MCF10A cells expressing variants H1047R, E545K, G1049R and other activating PIK3CA alleles (Q546K, N345K, H1047L, E542K) were significantly more sensitive to PI3K pathway inhibition (PI3K/mTOR inhibitor, BEZ235; PI3Kα inhibitor, BYL719; AKT inhibitor, MK2206) compared to parental cells and cells expressing wild-type or PIK3CAT1025T (Fig. 4B). These data suggest that patients whose tumors harbor rare, activating PIK3CA variants may benefit from therapeutic intervention similar to those carrying hotspot mutations.
Figure 4. PIK3CA tail mutations differentially activate cancer signaling pathways and sensitize cells to pathway inhibitors.
(A) Immunoblot analysis of the indicated proteins using extracts from MCF10A cells expressing wild-type (WT), rare (green) and hotspot (red) PIK3CA variants grown in the absence of insulin and EGF. Vinculin = loading control. (B) Dose-response survival curves for the indicated MCF10A stable cell lines treated with PI3K pathway inhibitors BEZ235, MK2206 and BYL719. (C) Representative differentially expressed proteins (p<0.05) determined by RPPA, respectively, from the indicated stable MCF10A cell lines grown in the absence of insulin and EGF. Entire RPPA dataset (p<0.05) provided in Supplementary Figure S2. (D) Dose-response survival curves for the indicated MCF10A stable cell lines treated with HER2/EGFR inhibitors Lapatinib and Neratinib. (E) Immunoblot analysis of the indicated proteins using extracts from MCF10A cells expressing wild-type (WT), rare (green) and hotspot (red) PIK3CA variants grown in the absence of insulin and EGF. Vinculin = loading control. (F) Dose-response survival curves for the indicated MCF10A (left) and Ba/F3 (right) stable cell lines treated with inhibitors BEZ235, MK2206 and BYL719. For Ba/F3, the sensitivity exhibited by experimental cell lines (-IL3) is assessed by comparison with cell lines re-addicted to IL3 (+IL3) over PIK3CA variants. Error bars depict the standard deviation.
To expand our analyses of pathways activated by PIK3CA variant clones, we next applied Reverse Protein Phase Array (RPPA) technology (33) to MCF10A cell lysates to catalog PIK3CA effects on expression or phosphorylation of 214 cancer-related proteins. In total, we analyzed 32 lysates extracted from growth factor-restricted MCF10A cells expressing all 24 missense mutation variants and 8 controls (parental, 2x eGFP, wild-type PIK3CA, and 4 silent mutations). RPPA analysis revealed that expression of activating PIK3CA variants leads to a significant shift in diverse cancer-related pathways beyond changes directly associated with PI3K/AKT/mTOR signaling (Fig. 4C and Supplementary Fig. S2), a finding consistent with a recent global analysis of gene expression and metabolomics of PIK3CAH1047R-expressing MCF10A cells (34). RPPA confirmed significant activation of the PI3K/AKT/mTOR signaling axis and downstream effectors (Fig. 4C; p<0.05). In addition, activating PIK3CA variant-expressing cells exhibited robust phosphorylation or up-regulation of cell cycle proteins (e.g., phospho-Rb, CyclinB1 and FOXM1) and down-regulation of the p27KIP1 cell cycle inhibitor, phospho-ATM(S1981), phospho-Chk1(S345) and phospho-Chk2(T68), which are indicative of evasion of growth suppression and activated replication, both hallmarks of cancer consistent with the known role of PI3K pathway in cancer (7,35–38).
In addition to PI3K signaling, RPPA analysis also revealed significant (p<0.05) up-regulation of phospho-HER2 and phospho-EGFR (Fig. 4C and Supplementary Fig. S2), which are receptor tyrosine kinases (RTKs) positioned upstream of PI3K and often targeted for activation in cancer. Our finding of HER2 and EGFR activation suggests the presence of positive feedback loop, indicating these RTKs may represent therapeutic liabilities in PIK3CA-activated cells. To examine this further, we treated PIK3CA-expressing MCF10A cell lines with HER2 and EFGR inhibitors Lapatinib and Neratinib. Consistent with our signaling observations made by RPPA, MCF10A cells expressing activated PIK3CA variants were significantly more sensitive to inhibition compared to parental cells and cells expressing wild-type or PIK3CAT1025T(Fig. 4D). RPPA analysis also indicated PIK3CA-dependent MEK1/2 activation (phosphorylation) and down-regulation of p38 and JNK (Fig. 4C and Supplementary Fig. S2; p<0.05), which was also confirmed by immunoblotting analysis with available antibodies (Fig. 4E). Therefore, we performed a similar drug sensitivity screen as in Figure 4B using Trametinib, a potent inhibitor of the MEK1/2 positioned in the mitogen activated protein kinase (MAPK) pathway (39). Unlike treatment with p38 and JNK inhibitors VX-702 and JNK-IN-8 (40,41), respectively, Trametinib inhibited cell growth by MCF10A cells expressing activated PIK3CA variants H1047R, E545K, G1049R, Q546K, N345K, H1047L, E542K) compared to parental cells and cells expressing wild-type or PIK3CAT1025T (Fig. 4F). We confirmed these results using a subset of activating PIK3CA variants (H1047R, Q546K, G1049R, N345K, E545K) expressed in Ba/F3 under conditions of PIK3CA addiction (-IL3), and parallel counter screens on these cells re-addicted to IL3 in place of PIK3CA relieved the inhibitory growth effect by Trametinib (Fig. 4F). These data suggest crosstalk between PI3K and MAPK MEK1/2 module but not the p38/JNK module of MAPK pathway, which has been suggested elsewhere (42).
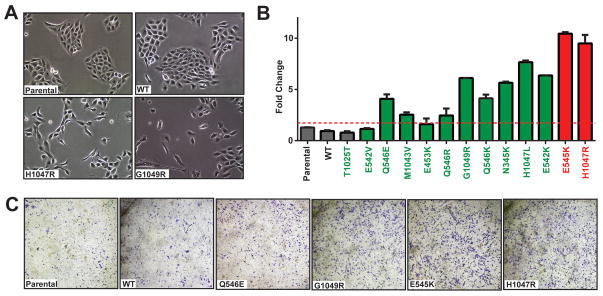
A previous study reported a role for PIK3CAH1047R in promoting epithelial-mesenchymal transition (EMT) and cell invasion, an obligate step during tumor metastasis, through activation of PI3K pathway (43). Consistent with this report, overexpression of activated PIK3CA variants led to striking morphological changes such as a cell spreading phenotype that often correlates with invasiveness, disrupting the typical “cobblestone” appearance of primary epithelial cells such as MCF10A (Fig. 5A). Moreover, our RPPA analysis (Fig. 4C, Supplementary Figure S2) revealed a significant increase in the expression of Rab25, Claudin-7, Caveolin and phospho-Myosin-IIa reported by others to represent molecular markers of cell invasion (44–47). To determine whether expression of PIK3CA variants is sufficient to promote cell invasion, we examined the invasiveness of MCF10A cells using transmembrane chambers coated with Matrigel (BD Biosciences). Stable expression of both PIK3CAH1047R and PIK3CAE545K hotspot variants led to a robust increase in cell invasion (9.6- and 10.6-fold, respectively, p<0.0001) compared to parental cells and those expressing wild-type or PIK3CAT1025T (Fig. 5B–C). PIK3CAE542V-and PIK3CAE453K-expressing cells did not provide pro-invasion activity consistent with their weak driver activity across the other phenotyping platforms used (Figs. 2–3) whereas the remaining 7 variants examined enhanced invasion at range of 2.5- to 7.8-fold over the mean of negative controls (Fig. 5B) in a manner consistent with signaling assessed by RPPA (Fig. 4C).
Figure 5. Oncogenic PIK3CA variants induce invasive phenotypes in breast epithelial cells.
(A) Representative images of parental MCF10A cells and those expressing wild-type (WT) or the indicated PIK3CA variants (PIK3CAH1047 and PIK3CAG1049R). (B) MCF10A cells expressing the indicated controls, wild-type (WT), low-frequency (green) and hotspot (red) PIK3CA variants were applied to transwell Matrigel invasion chambers, followed by quantitation of invaded cells. Error bars depict the standard deviation. (C) Representative images from bottoms of cell invasion chambers from (B) following staining with crystal violet.
DISCUSSION
Recent developments in NGS technology are enabling full cataloguing of genomic alterations residing within cancer genomes at record pace and lower costs, revealing numerous potential targets and therapeutic approaches for the treatment of cancer. However, translation of NGS-based discoveries is hampered by the slower development of functional genomics approaches aimed at characterizing newly discovered “uncommon” aberrations and their impact on therapeutic response. In this study, we attempted to narrow this gap by designing scalable screening tools for constructing and examining rare gene variants. Our comprehensive mutation assessment platform integrates an in-house developed, high-throughput mutagenesis and barcoding technology with a phenotype screening to rapidly interrogate each variant for cancer-relevant activities. Our results provided key insights on correlation between rare events of PIK3CA and their oncogenic potentials as well as novel implications for cancer therapy. The scalability and success of our approaches toward discovering rare, activating cancer mutations suggests this platform may applied to other cancer types to screen for novel cancer-causing aberrations following tissue-specific modifications of the models employed.
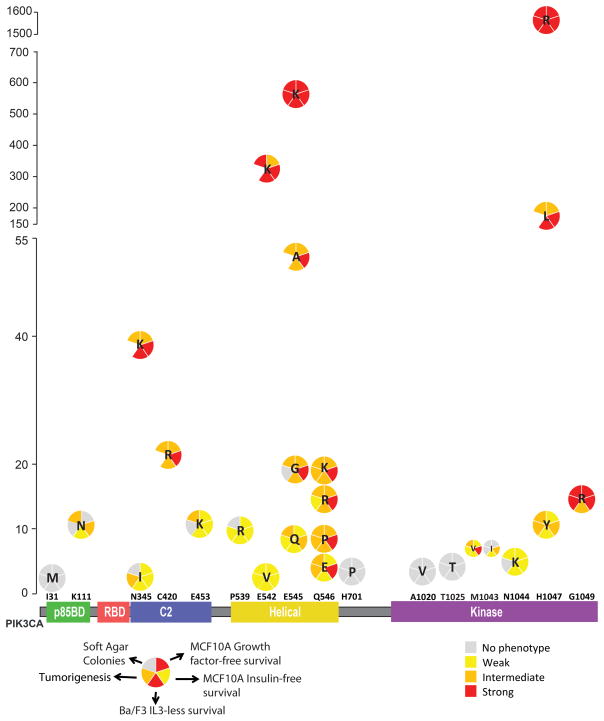
We first used our platform to examine an allelic series of the PIK3CA gene, a well-known oncogene often targeted by mutagenesis in cancer. Some attempts have been made to catalogue the consequences of expressing rare PIK3CA mutant alleles [e.g., (8–11). However, such studies employ varying expression systems and diverse cell models that lack tissue specificity for the study of PIK3CA variants identified in breast cancer, which collectively make it difficult to compare PIK3CA variant driver activity across those studies. In addition to Ba/F3 cells, which provide a generalized “sensor” model to detect driver activity, we also employed normal MCF10A breast cells as well as primary HMECs to provide lineage specificity. Our results revealed that 21 out of 25 (84%) rare PIK3CA mutations (COSMIC frequencies ranging <0.1–10%) were oncogenic. Interestingly, the power of PIK3CA variant activity correlated significantly with their frequency in breast cancer, albeit not perfectly (Fig. 6). Of note, mutations that were present on the same residues had different phenotypic activity correlated with their frequencies. While higher frequency mutation on a given residue showed a stronger phenotype, rare mutations of the same residue exhibited weaker activity (see Fig. 6 for mutations of N345, E542, E545, Q546 and H1047). Q546 mutations were all similar to each other in terms of activity in our phenotyping assays, which is consistent with their comparable frequencies. One intriguing result from our screening platform was that even though G1049R mutation is a rare (~0.5%) aberration, it demonstrated strong driver activity in 4 out of 5 assays used in our platform and exhibited activity levels similar to the E542K variant with 20-fold higher frequency (10.8%; Fig. 6). Further studies need to address why this strong event is present at low frequencies in breast cancer.
Figure 6. PIK3CA tail mutation activity summary.
X-axis represents the domain structure of PIK3CA protein (p110alpha) with analyzed mutations depicted (x-axis is not to scale). Y-axis represents the occurrence of each mutation based on COSMIC analysis. Relative strength of phenotype for each assay is based on as follows: Ba/F3: Weak=2–15 fold, Intermediate=15–35 fold, Strong>35 fold; Growth factor-free proliferation: Weak=2–4 fold, Intermediate=4–9 fold, Strong>9 fold; No Insulin proliferation: Weak=1.3–2.5 fold, Intermediate=2.5–4 fold, Strong>4 fold and above; Colony formation: Weak=25–35 colonies, Intermediate=35–65 colonies, Strong=>65 colonies; Tumorigenesis: Weak=1–5% representation, Intermediate=5–10% representation, Strong>10% representation.
Deconvoluting the alterations at a molecular level triggered by expression of PIK3CA variants revealed expected consequences as well as novel deregulation of cancer pathways. Consistent with the previous data, oncogenic PIK3CA variants also deregulated transcription and translation pathways and led to activated cell cycle and down-regulation of stress/inflammation markers (48) in accordance with mechanistic alterations caused by other oncogenes. Proteomics analysis and our observation of phenotypic changes in 2-D cultures of PIK3CA variant-expressing cells also suggested that oncogenic PIK3CA alleles drive invasiveness, which was confirmed with increased invasion potential in transwell chambers. In addition, active PIK3CA mutations hyper-activated PI3K pathway and rendered cells sensitive to inhibitors targeting PI3K pathway as expected. However, we also observed activation of MAPK pathway, a commonly deregulated growth regulating cancer pathway, suggesting a strong positive cross-talk between PI3K and MAPK signaling. PIK3CA variants led to hyper-activation of EGFR, HER2 receptors and MEK1/2 modulators, and as a result, cells expressing PIK3CA variants were sensitized to inhibitors of these already known cancer targets. Our findings imply that patients that are positive for PIK3CA variants might benefit from combinatorial treatment options targeting both PI3K and MAPK pathways.
It has been suggested before that based on the affected domains, PIK3CA variants induce gain-of-function by different mechanisms (49–51). Our results in this study informed us on alterations and therapeutic liabilities that are common to all active PIK3CA variants; however, as a next step, it would be intriguing to study the potential mechanistic differences between PIK3CA alleles categorizing them into different classes with their corresponding unique drug liabilities. This would take precision medicine one step further from “gene specific” targeting to “mutation specific” approach in hopes of more efficient therapeutic options.
Supplementary Material
Acknowledgments
GRANT SUPPORT
This project was supported in part by the Genomic and RNA Profiling Core at Baylor College of Medicine with funding from the NIH/NCI grant (P30CA125123). This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT; RP120046) by funding to K.L.S. This work was also supported by the NIH (UO1CA168394) by funding to K.L.S and G.B.M. T.D. was supported by a training grant from The Cullen Foundation. Further support was obtained from the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy and The Bosarge Family Foundation.
Footnotes
Disclosure of potential conflicts of interest: No potential conflicts of interest were disclosed.
AUTHORS’ CONTRIBUTIONS
Conception and design: T. Dogruluk, K.L. Scott
Development of methodology: T. Dogruluk, K.L. Scott
Acquisition of data (provided animals, provided facilities, etc.): T. Dogruluk, Y.H. Tsang, M. Espitia, A. Dogruluk, K. Eterovic, G.B. Mills, K.L. Scott
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): T. Dogruluk, F. Chen, T. Chen, Z. Chong, V. Appadurai, P.E. Bonnen, C.J. Creighton, K. Chen, G.B. Mills, K.L. Scott
Writing, review and/or revision of the manuscript: T. Dogruluk, K. Eterovic, P.E. Bonnen, C.J. Creighton, K. Chen, G.B. Mills, K.L. Scott
Administrative, material, or technical support (i.e., reporting data or organizing data, constructing databases): T. Dogruluk, T. Chen, Z. Chong, K. Chen, K.L. Scott
Study supervision: K.L. Scott
References
- 1.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7(6):561–73. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65(23):10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 6.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102(3):802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 8.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007;104(13):5569–74. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65(11):4562–7. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 10.Hon WC, Berndt A, Williams RL. Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases. Oncogene. 2012;31(32):3655–66. doi: 10.1038/onc.2011.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA) Proc Natl Acad Sci U S A. 2012;109(38):15259–64. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung LW, Yu S, Zhang D, Li J, Ng PK, Panupinthu N, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell. 2014;26(4):479–94. doi: 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 16.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong WC, Kim D, Carter H, Diekhans M, Ryan MC, Karchin R. CHASM and SNVBox: toolkit for detecting biologically important single nucleotide mutations in cancer. Bioinformatics. 2011;27(15):2147–8. doi: 10.1093/bioinformatics/btr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–2. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88(4):440–9. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao Y, Chen H, Liang H, Meric-Bernstam F, Mills GB, Chen K. CanDrA: cancer-specific driver missense mutation annotation with optimized features. PLoS One. 2013;8(10):e77945. doi: 10.1371/journal.pone.0077945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung LWT, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, et al. High Frequency of PIK3R1 and PIK3R2 Mutations in Endometrial Cancer Elucidates a Novel Mechanism for Regulation of PTEN Protein Stability. Cancer Discovery. 2011;1(2):170–85. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kammann M, Laufs J, Schell J, Gronenborn B. Rapid insertional mutagenesis of DNA by polymerase chain reaction (PCR) Nucleic Acids Res. 1989;17(13):5404. doi: 10.1093/nar/17.13.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wardwell-Ozgo J, Dogruluk T, Gifford A, Zhang Y, Heffernan TP, van Doorn R, et al. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene. 2014;33(8):1017–26. doi: 10.1038/onc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu W, Jia Z, Liu P, Xie Z, Wei Q. A novel PCR strategy for high-efficiency, automated site-directed mutagenesis. Nucleic Acids Res. 2005;33(13):e110. doi: 10.1093/nar/gni115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41(3):727–34. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 28.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22(11):2120–9. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin SI, Freedman VH, Risser R, Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci U S A. 1975;72(11):4435–9. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15(1):50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollier BG, Tinnirello AA, Werden SJ, Evans KW, Taube JH, Sarkar TR, et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013;73(6):1981–92. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clinical proteomics. 2010;6(4):129–51. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hart JR, Zhang Y, Liao L, Ueno L, Du L, Jonkers M, et al. The butterfly effect in cancer: a single base mutation can remodel the cell. Proc Natl Acad Sci U S A. 2015;112(4):1131–6. doi: 10.1073/pnas.1424012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita N, Sato S, Tsuruo T. Phosphorylation of p27Kip1 at threonine 198 by p90 ribosomal protein S6 kinases promotes its binding to 14-3-3 and cytoplasmic localization. J Biol Chem. 2003;278(49):49254–60. doi: 10.1074/jbc.M306614200. [DOI] [PubMed] [Google Scholar]
- 36.Shin I, Yakes FM, Rojo F, Shin NY, Bakin AV, Baselga J, et al. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med. 2002;8(10):1145–52. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- 37.Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3(3):245–52. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 38.Park HJ, Carr JR, Wang Z, Nogueira V, Hay N, Tyner AL, et al. FoxM1, a critical regulator of oxidative stress during oncogenesis. Embo J. 2009;28(19):2908–18. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res. 2011;17(5):989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 40.Lee MR, Dominguez C. MAP kinase p38 inhibitors: clinical results and an intimate look at their interactions with p38alpha protein. Curr Med Chem. 2005;12(25):2979–94. doi: 10.2174/092986705774462914. [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, Inesta-Vaquera F, Niepel M, Zhang J, Ficarro SB, Machleidt T, et al. Discovery of potent and selective covalent inhibitors of JNK. Chemistry & biology. 2012;19(1):140–54. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ebi H, Costa C, Faber AC, Nishtala M, Kotani H, Juric D, et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc Natl Acad Sci U S A. 2013;110(52):21124–9. doi: 10.1073/pnas.1314124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallin JJ, Guan J, Edgar KA, Zhou W, Francis R, Torres AC, et al. Active PI3K pathway causes an invasive phenotype which can be reversed or promoted by blocking the pathway at divergent nodes. PLoS One. 2012;7(5):e36402. doi: 10.1371/journal.pone.0036402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joglekar M, Elbazanti WO, Weitzman MD, Lehman HL, van Golen KL. Caveolin-1 Mediates Inflammatory Breast Cancer Cell Invasion via the Akt1 Pathway and RhoC GTPase. J Cell Biochem. 2015;116(6):923–33. doi: 10.1002/jcb.25025. [DOI] [PubMed] [Google Scholar]
- 45.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18(8):3144–55. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahiya N, Becker KG, Wood WH, 3rd, Zhang Y, Morin PJ. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS One. 2011;6(7):e22119. doi: 10.1371/journal.pone.0022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amornphimoltham P, Rechache K, Thompson J, Masedunskas A, Leelahavanichkul K, Patel V, et al. Rab25 regulates invasion and metastasis in head and neck cancer. Clin Cancer Res. 2013;19(6):1375–88. doi: 10.1158/1078-0432.CCR-12-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5(12):921–9. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 49.Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, et al. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J. 2008;409(2):519–24. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 50.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317(5835):239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 51.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105(7):2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.