Abstract
Background
MicroRNAs are key players in cardiac stress responses, but the mRNAs whose abundance and/or translational potential are primarily affected by changes in cardiac microRNAs are not well defined. Stimulus-induced, large-scale alterations in the cardiac transcriptome, together with consideration of the law of mass action, further suggest that the mRNAs most substantively targeted by individual microRNAs will vary between unstressed and stressed conditions. To test the hypothesis that microRNA target profiles differ in health and disease, we traced the fate of empirically-determined miR-133a and miR-378 targets in mouse hearts undergoing pressure-overload hypertrophy.
Methods and Results
Ago2 immunoprecipitation with RNA-sequencing (RISC-sequencing) was used for unbiased definition of microRNA-dependent and –independent alterations occurring amongst ~13,000 mRNAs in response to transverse aortic constriction (TAC). Of 37 direct targets of miR-133a defined in unstressed hearts (fold-change ≥25%, FDR<0.02), only 4 (11%) continued to be targeted by miR-133a during TAC, while for miR-378 direct targets, 3 of 32 targets (9%) were maintained during TAC. Similarly, only 16% (for miR-133a) and 53% (for miR-378) of hundreds of indirectly affected mRNAs underwent comparable regulation, demonstrating that the effect of TAC on microRNA direct target selection resulted in widespread alterations of signaling function. Numerous microRNA-mediated regulatory events occurring exclusively during pressure overload revealed signaling networks that may be responsive to the endogenous decreases in miR-133a during TAC.
Conclusions
Pressure overload-mediated changes in overall cardiac RNA content alter microRNA targeting profiles, reinforcing the need to define microRNA targets in tissue-, cell- and status-specific contexts.
Keywords: miR, microRNA, pressure overload, gene expression/regulation, mRNA, Ago2, RNA-induced silencing complex, microRNA binding protein, Argonaute 2
Introduction
MicroRNAs are short noncoding RNAs that serve to limit the translation of specific mRNAs, often but not always observed in conjunction with mRNA transcript degradation1, 2. MicroRNAs bound to Argonaute proteins recruit other protein components of RNA-induced silencing complexes (RISCs) to specific sites on target mRNAs. One key determinant of the strength of the mRNA-RISC interaction is the extent of nucleotide hybridization in the region of the initially exposed microRNA nt 2–7 ‘seed’ region3. Additional important parameters include the nucleation free energy of the microRNA-mRNA interaction4, the accessibility of putative mRNA target regions, and conformational constraints placed on microRNAs by Argonaute binding5. Furthermore, as for other biomolecular interactions, the amount of RISC-targeted mRNA depends not only on binding affinities but also on the concentrations of both the mRNA and of appropriately microRNA-programmed RISCs (i.e. the law of mass action). Such data are critical to determining the final extent of interactions between individual microRNAs and mRNAs in biological contexts, given the degenerate nature of microRNA-mRNA interactions and the likelihood that numerous mRNAs will compete for binding to the same microRNA.
The contribution of microRNAs to pathological cardiac remodeling induced by pressure overload has been studied by many groups, with key roles suggested for miR-16–8, -34a9, -15510, -199b11, -208a12, and -212/13213 among others. Despite potent effects of microRNA inhibition or elevation on hypertrophic outcomes, relatively few experimentally validated mRNA targets of these microRNAs have been uncovered. In addition, mRNA targets of microRNAs are frequently defined from in vitro models or unstressed hearts and assumed to continue to represent substantive targets of these microRNAs in the disease state. Considering that stressors provoke widespread changes in the cardiac cohort of microRNAs and mRNAs14, 15, thus altering the concentrations of the possible reactants in mRNA-RISC associations, we hypothesized that the mRNAs most substantially targeted by a given microRNA may differ between unstressed and stressed hearts. The significance of this hypothesis is that, if supported, proper understanding of microRNA-dependent contributions to signaling pathways and planned microRNA-based interventions in pathological circumstances would need to be based primarily on RNA interaction data obtained from hearts exhibiting that same pathology, rather than from other models.
To test these hypothesis, we took advantage of existing mice with cardiac-specific overexpression of two abundant microRNAs of importance to the heart, miR-133a and miR-378. We have previously developed methods (RISC-sequencing) to define mRNAs that undergo increased targeting by RISCs in response to cardiac microRNA overexpression in vivo14, 16–18. While the endogenous levels of these two microRNAs are decreased during transverse aortic constriction (TAC)19–24, persistent microRNA overexpression in both non-diseased and diseased states rendered comparisons of targeting profiles before and after TAC possible. By tracking the behavior of separate cohorts of microRNA-dependent mRNAs before and after TAC, we demonstrate here that the precise RISC-mediated mRNA suppression events that take place in response to either transgene are substantially altered under TAC conditions.
Methods
Mouse microRNAs are described according to the nomenclature used by miRBase 21, released in June 2014 (http://www.mirbase.org/)25, 26; ‘miR-133a’ refers to mmu-miR-133a-3p and ‘miR-378’ refers to mmu-miR-378a-3p unless otherwise mentioned.
Mice overexpressing pre-miR-133a and pre-miR-378 under the control of the Myh6 promoter (αMHC-miR-133a and αMHC-miR-378 mice) were generated as previously described21, 27. All procedures used male mice, 8–12 weeks of age and were performed in accordance with the policies of the Animal Studies Committee at Washington University School of Medicine. For miR-133a studies, RNA-sequencing libraries were prepared from 3 nontransgenic littermate controls and 7 αMHC-miR-133a mice subjected to sham surgery, and 5 nontransgenic and 4 αMHC-miR-133a mice subjected to transverse aortic constriction (TAC). For miR-378 studies, libraries were prepared from 3 nontransgenic and 3 αMHC-miR-378 mice subjected to sham surgery, and 7 mice of each genotype subjected to TAC.
Unless otherwise sp, regulated RNAs were defined using a threshold of 25% (increased or decreased) at a false discovery rate (FDR) of 0.02.
All mRNA-seq and RISC-seq data from this study have been deposited in the NCBI GEO under accession GSE65141 (for the miR-133a study cohort) and GSE61734 (for the miR-378 study cohort).
Detailed protocols and procedures for all experiments (Expanded Methods) are in the Supplemental Material.
Results
RISC-bound RNA and global mRNA alterations in response to pressure overload
Previous studies from our group have documented dysregulation of microRNAs and mRNAs and consequent effects on microRNA suppression of mRNAs during murine cardiac hypertrophy induced by pressure overload14. Our earlier investigation focused especially on TAC regulation of a) mRNAs that were already substantively suppressed (enriched in RISCs compared to global mRNA) under sham conditions, and b) which exhibited converse regulation in the RISC-bound and global mRNA fractions14. However, accumulating studies demonstrate that alterations in mRNA-RISC association manifest as changes in mRNA translation (ribosome initiation and elongation and ultimately protein content) that are temporally dissociated from changes in mRNA transcript levels2, 28. Thus, we analyzed data from a new cohort of mice subjected to 1 week of pressure overload and added a further category c); alterations in mRNA RISC abundance without alterations in global mRNAs, considered as transcript level-independent changes in mRNA translational competence (Expanded Methods). While the above definition captures a wider set of RISC-dependent mRNAs than that presented in prior studies14, 16, 17, protein functional subcategories of transcription factors, kinases and phosphatases continued to be overrepresented in comparisons of microRNA-dependent mRNAs defined in this manner compared to microRNA-independent mRNAs (Supplemental Table 1), similar to previous findings14.
The RISC-enrichment ratio (or RISC score) describes the amount of an individual mRNA measured in sequencing libraries prepared from Ago2 immunoprecipitates (Figure 1a), compared to the amount of the same mRNA measured in libraries prepared from total cellular mRNA14, 16, 17 (Figure 1b). The mRNAs affected by TAC in a microRNA-dependent manner were not exclusively mRNAs with high RISC scores in sham-operated mice, nor were they exclusively mRNAs that tended not to be microRNA-targeted under sham conditions (Figure 1b, Supplemental Table 2). Together with the 911 mRNAs directly altered in a microRNA-dependent manner by TAC, an additional 1,766 mRNAs were upregulated and 1,948 were downregulated without RISC changes (Supplemental Table 3), suggesting that at least some direct mRNA targets of microRNAs feed back on nuclear signaling and promoter-driven mRNA transcription14, 17. Several previously described targets of TAC-regulated microRNAs from the literature were evaluated for changes in RISC RNA and total mRNA abundance (Figure 2). Strikingly, many of these mRNAs did not undergo alterations during TAC that would have been predicted from changes in their upstream microRNAs alone, revealing the importance of additional factors in determining final mRNA abundances and translational outcome.
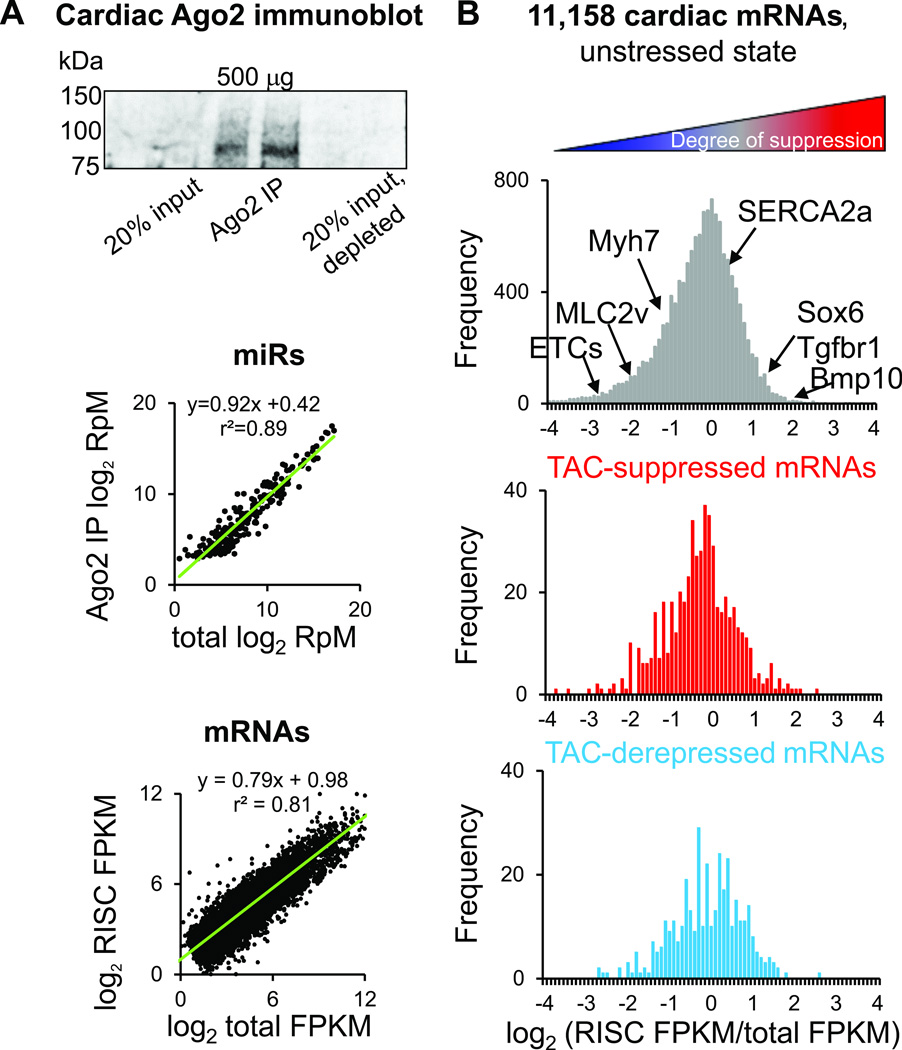
Figure 1.
Effect of pressure overload on RISC-bound and global mRNAs. A) Top, representative Ago2 immunoprecipitation from mouse heart; center, global microRNA vs RISC-bound (Ago2-immunoprecipitated) microRNA abundances for 300 detected cardiac microRNAs14; bottom, global mRNA vs RISC-bound (Ago2-immunoprecipitated) mRNA abundances for 11,158 detected cardiac mRNAs in nontransgenic, unstressed mice. RpM, reads per million mapped reads (microRNAs); FPKM, fragments per exon of transcript per million mapped reads (mRNAs). B) Histogram of log2-transformed RISC ratios (abundance in RISC / abundance in global mRNA) in nontransgenic, sham-operated mice. Lower panels show log2 ratio positions of mRNAs suppressed or derepressed in a microRNA-dependent manner after 1 week TAC (FDR<0.02).
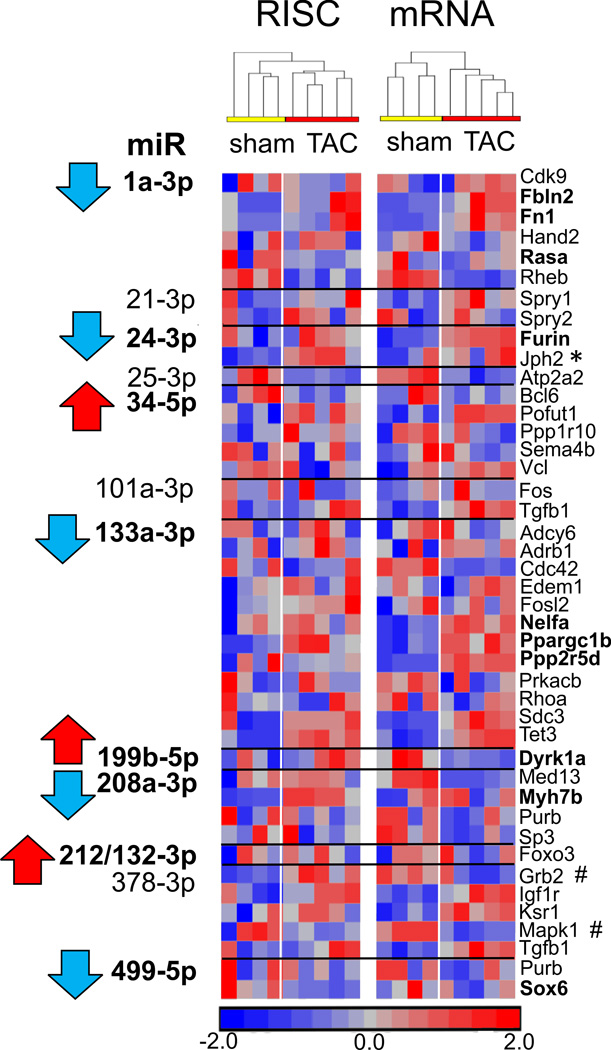
Figure 2.
Effect of pressure overload on previously described mRNA targets of regulated microRNAs. RISC-bound and global mRNA levels from 1 wk TAC hearts are shown in a standardized heatmap, for microRNA-mRNA regulatory pairs described in previous reports. Red = increased mRNA abundance in RISC-bound or global fractions, blue = decreased RNA abundance. MicroRNAs observed to be regulated at 1 wk TAC in our studies14 are shown in bold, and mRNAs that exhibit predicted behavior are also shown in bold. * denotes that mRNA exhibits behavior in TAC consistent with previous report(s) but proposed regulatory microRNA does not change in the same way (e.g. miR-24-3p, Jph2). # denotes that mRNA exhibits transcript abundance consistent with previous reports in TAC, but does not appear to be dependent on proposed regulatory microRNA (Grb2, Mapk1). mRNA targets of microRNAs are drawn from the following studies: miR-1a-3p6, 7, 29; miR-21-3p30, 31; miR-24-3p32, 33; miR-25-3p34; miR-34-5p family9, 35; miR-101a-3p36; miR-133a-3p16, 20, 22; miR-199b-5p11; miR-208a-3p37, 38; miR-212/132-3p13; miR-378-3p23, 24, 39; miR-499-5p38.
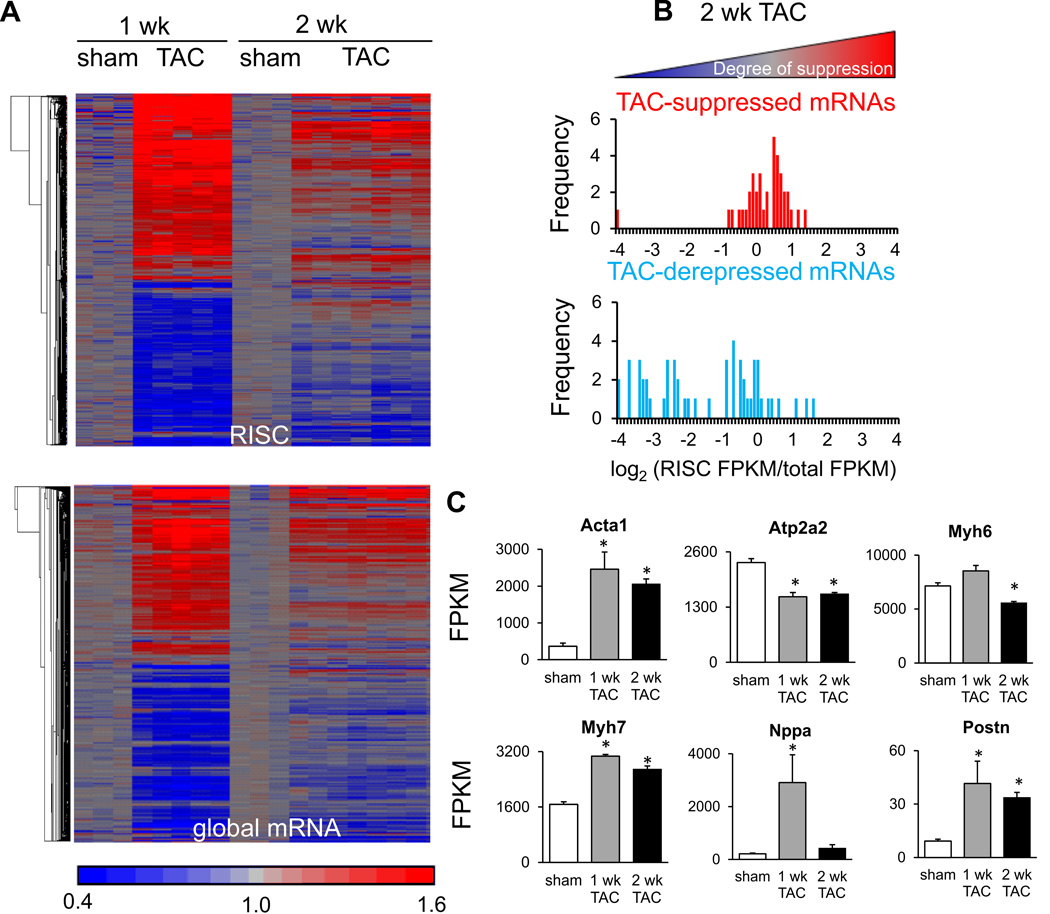
To assess whether a majority of microRNA-dependent regulatory events had been captured by examining hearts after 1 week of TAC, and to examine whether these events were maintained, further assays were performed at a 2-week timepoint. Pressure overload induces an initial concentric hypertrophy which then progresses toward chamber dilation40; echocardiographic data from the hearts used for RNA assays demonstrated a decreased r/h ratio at 1 week but not at 2 weeks (Supplemental Figure 1). Comparison of both RISC-level and total mRNA-level alterations at these time points demonstrated much less pronounced microRNA-dependent regulation after 2 weeks of TAC (Figure 3a, 3b), although typical changes in hypertrophic ‘marker’ mRNAs selected from those altered during the fetal-adult transition were observed over the same time frame (Figure 3c). These data indicate that the acute response to pressure overload is not sustained, but rather that changes to the transcriptional and translational program occur later in TAC.
Figure 3.
Effect of sustained pressure overload on RISC-bound and global mRNAs. A) ~2300 regulated mRNAs (FDR<0.02) in either RISC-bound (upper panel) or global mRNA fractions (lower panel) at either 1 week or 2 week TAC, displayed as standardized heatmaps. Each column represents an individual heart; signals were transformed such that the mean value for sham-operated hearts is equal to 1. Row order of mRNAs in the upper panel is not the same as in the lower panel. 3 sham and 5 TAC hearts are shown for the 1 week condition; 3 sham and 7 TAC hearts are shown for the 2 week condition. B) MicroRNA-dependent, suppressed and derepressed mRNAs shown according to unstressed RISC ratio values (similarly to Figure 1b) but for 2 week TAC (FDR<0.02). C) Typical mRNA markers of cardiac hypertrophy evaluated at 1 and 2 week TAC via mRNA-sequencing (* = FDR<0.02 compared to sham); FPKM, fragments per exon of transcript per million mapped reads.
Cardiomyocyte and nonmyocyte contribution to TAC-regulated mRNAs
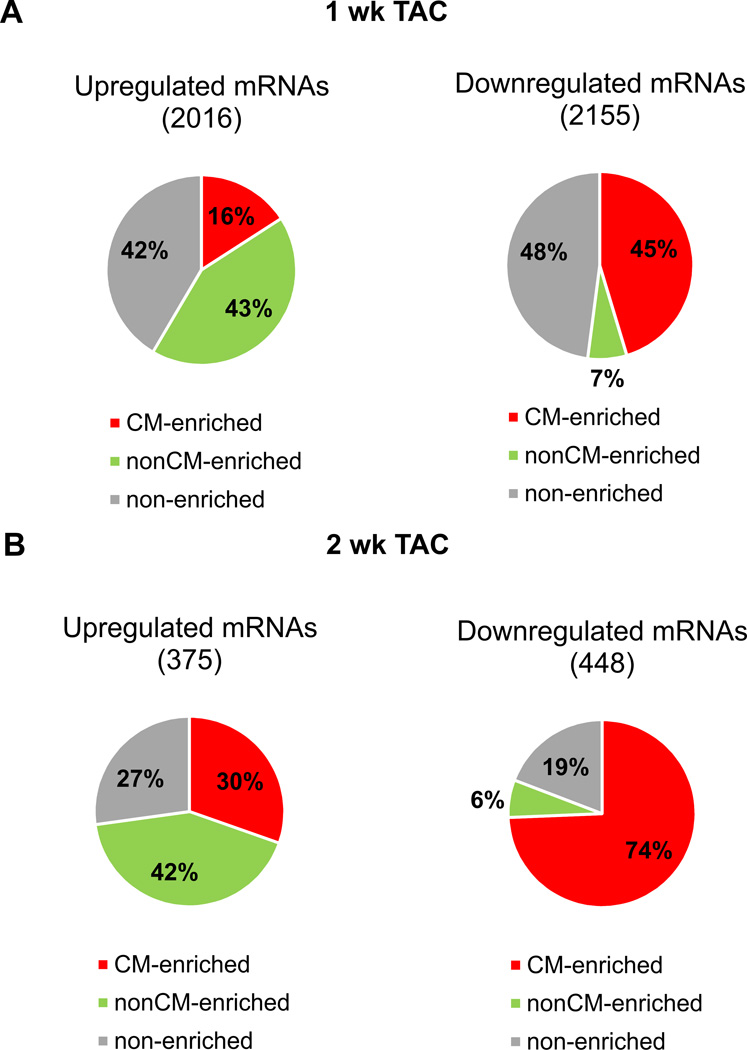
In preparation for studies intended to monitor the behavior of cardiomyocyte mRNAs targeted by microRNAs during TAC, we first established which regulatory events were most likely taking place in cardiomyocytes. We took advantage of isolated adult cardiomyocyte and nonmyocyte microRNA- and mRNA-sequencing profiles obtained during previous studies27, 41 and classified regulated RNAs from the 1- and 2-week TAC studies according to enrichment in myocyte or nonmyocyte fractions (Expanded Methods). Most mRNAs (and microRNAs) regulated by TAC fell into cardiomyocyte-enriched or cell type non-enriched categories (Figure 4, Supplemental Figure 2, Supplemental Tables 4 and 5). Notably, we found that downregulation was much more prominent among cardiomyocyte-enriched mRNAs, while upregulation occurred much more often for nonmyocyte-enriched mRNAs. This observation was made both at 1- and 2-week time points (Figure 4). Indeed, immunostaining and FACS studies have shown that the proportion of fibroblasts to cardiomyocytes markedly increases in early pressure-overload hypertrophy, such that these cells are present in a 1:1 ratio at 1 and 2 weeks after TAC compared to the wild-type 1:2 fibroblast:cardiomyocyte ratio42. Thus, we propose that at least some of the observed downregulation of cardiomyocyte-enriched RNAs and upregulation of nonmyocyte-enriched RNAs, when measuring global RNAs extracted from intact hearts, reflects an alteration in the cardiomyocyte:fibroblast mass ratio. These data also indicate that caution must be used when assigning likely in vivo mRNA targets of regulated microRNAs when the RNAs under consideration are not present in the same cellular enrichment fraction.
Figure 4.
Cardiomyocyte and nonmyocyte distribution of TAC-regulated mRNAs. A) Pie charts of global mRNAs regulated at FDR<0.02 after 1 week TAC. Slices in each pie show cardiomyocyte (CM)-enriched (red), nonCM-enriched (green), and non-cell type-enriched mRNAs (gray). B) as for A), but for 2 week TAC.
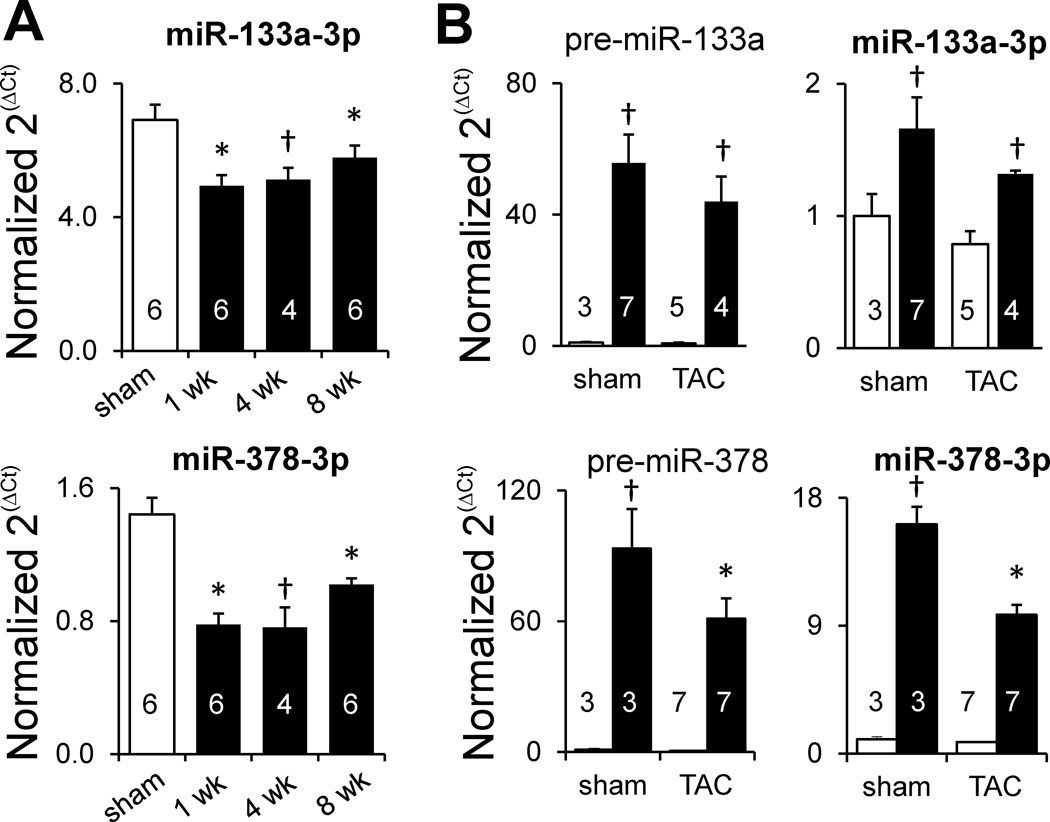
Identification of empirical mRNA targets of microRNAs resulting from microRNA overexpression
miR-133a and miR-378 are highly abundant cardiomyocyte-enriched microRNAs (Supplemental Figure 3) whose expression, when measured from intact heart RNA preparations, is persistently decreased in response to pressure overload (Figure 5a). We overexpressed each of these microRNAs in cardiomyocytes (Figure 5b) so that mRNA targets of the microRNA transgenes could be identified under unstressed conditions, and then could be further evaluated to judge whether these mRNAs were still regulated in a microRNA-dependent manner during TAC. Notably, a much higher degree of pre-miR overexpression was observed than that of the mature, processed microRNA, suggesting a bottleneck in downstream Dicer processing (Figure 5b). Using the same fold-change and statistical selection criteria as for the TAC studies in Figure 1, but also taking care to filter the data to include only cardiomyocyte-enriched (or non-cell type-enriched mRNAs), 183 mRNAs displayed increased (microRNA-dependent) RISC association in response to αMHC-miR-133a (Supplemental Table 6a). As might be expected from altered direct RISC targeting of several mRNAs, a consequence of miR-133a overexpression was RISC-independent (i.e. indirect) regulation of many further mRNAs (Supplemental Table 6b; 142 upregulated and 267 downregulated, excluding 86 regulated mRNAs enriched in the nonmyocyte fraction). The RNA-sequencing data on mRNA abundance changes in response to miR-133a compared favorably to previous microarray measurements of regulated transcripts in αMHC-miR-133a hearts21 (Supplemental Figure 4).
Figure 5.
Endogenous miR-133a and -378 decrease in TAC and transgene overexpression. A) RT-qPCR measurement of miR-133a and -378 during sustained TAC (normalized to U6 expression). White bars, sham; black bars, TAC. B) RT-qPCR measurement of microRNA precursors and mature microRNAs during 1 week TAC (for miR-133a) and 2 week TAC (for miR-378). White bars, nontransgenic; black bars, transgenic. * P<0.05 relative to sham (unpaired t-test); † P<0.05 relative to sham (Mann-Whitney). Numbers in bar plots designate biological replicates. Different mouse hearts were used in nontransgenic sham and TAC conditions shown in panel A to those used in panel B.
We observed similar increases and decreases in global mRNA abundances in αMHC-miR-378 hearts to those previously reported27 (309 upregulated and 361 downregulated, excluding 147 regulated mRNAs enriched in the nonmyocyte fraction), but relatively few mRNAs whose direct association with the RISC was increased in response to the transgene (only 7 mRNAs at FDR<0.02) (Supplemental Table 7). At a less stringent statistical cutoff (FDR<0.1), 38 mRNAs had increased RISC associations (Supplemental Table 7). Although we have previously validated that mRNAs without significant global abundance changes, but with significant increases in RISC association, are indeed subject to translational suppression17, we demonstrated that RISC-increased Gapdh is decreased at the protein level and that RISC-increased Rapgef4 is subject to miR-378 suppression in a luciferase reporter assay (Supplemental Figure 5).
Consistent with the notion that microRNA overexpression reprograms the RISC (and that the number of Ago2 molecules available for microRNA binding may represent a limiting factor), numerous mRNAs exhibited decreased rather than increased RISC abundance in the presence of either microRNA transgene (Supplemental Table 8). Such reprogramming could result from overexpressed microRNAs displacing other, less abundant microRNAs from available Ago2, and from secondary effects of microRNA overexpression that change levels of other microRNAs (as we previously demonstrated for miR-37827). Although it would be critical to identify the roles of such ‘RISC-derepressed’ mRNAs when considering final phenotypes arising from microRNA overexpression, we did not evaluate these further for the purpose of the current studies, which focus on pressure-overload effects on readily classifiable, direct targets of microRNAs (defined as those with enhanced RISC binding).
Pressure overload competes with microRNA overexpression for regulation of numerous mRNAs
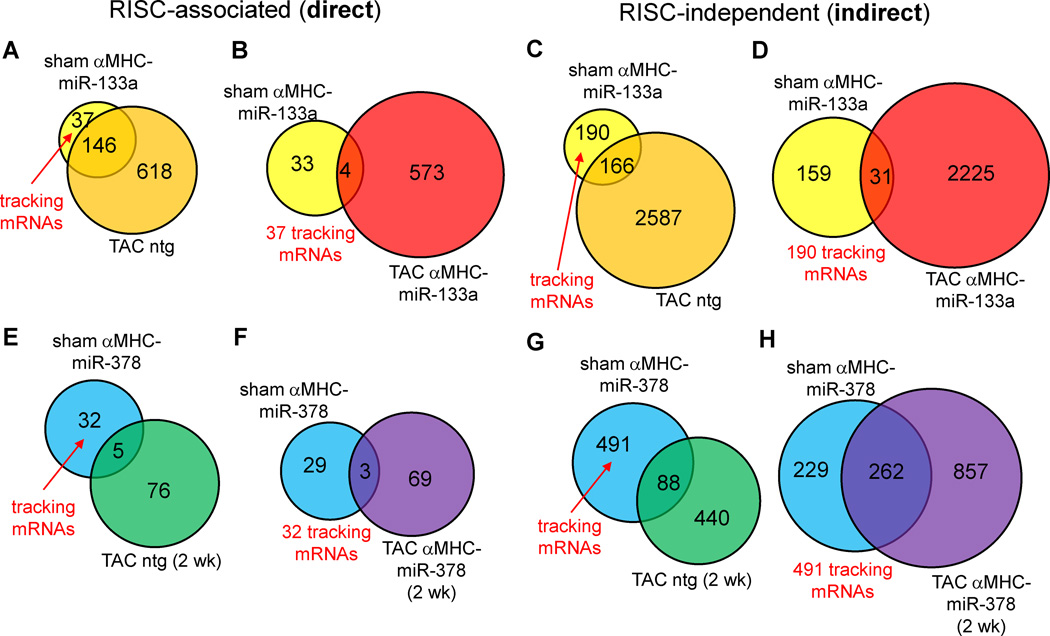
In order to be able to test the hypothesis that pressure overload-induced changes in RISC association profiles and mRNA abundances would alter functional microRNA targets, we needed to be able to track a set of microRNA-regulated mRNAs in the environment of TAC. Any independent effect of TAC (in nontransgenic mice) on any of the same mRNA targets identified in αMHC-miR hearts would confound further analyses. In addition, we were careful not to select ‘tracking’ mRNAs which appeared to be enriched in cardiac nonmyocytes, as these are less likely to be authentic direct mRNA targets of cardiomyocyte-overexpressed microRNAs. Of the starting 183 mRNAs directly targeted by miR-133a in unstressed hearts, 146 underwent regulation at the RISC and/or global mRNA level in response to TAC alone, leaving a set of 37 mRNAs that could be used to monitor the effect of TAC on miR-133a-dependent RISC association. These served as the miR-133a ‘tracking’ set (Supplemental Table 6c). Similarly, we established a set of 32 ‘tracking’ mRNAs directly targeted by miR-378 overexpression that were unaffected by TAC alone (Supplemental Table 7c). For αMHC-miR-378 mice and their nontransgenic controls, all RNA measurements were performed after 2 weeks of TAC (rather than the 1 week used for miR-133a studies) because parallel echocardiographic measurement suggested that this represented a key time point at which major functional differences were observed (vide infra).
We considered two broad classes of outcome with regard to RISC association of mRNAs in response to microRNA overexpression and TAC. Firstly, RISC programming in αMHC-miR hearts, and separate RISC programming and mRNA abundance changes caused by TAC-mediated changes in microRNA and mRNA transcription, could comprise largely independent, additive actions. Secondly, competition between microRNA transgene-programmed RISCs and TAC for overlapping sets of mRNAs could lead to a) synergistic effects from both stimuli; and b) events mediated by one stimulus that are canceled out by the other. Only 4 of the 37 mRNAs in the miR-133a-dependent ‘tracking set’ (11%) continued to exhibit increased RISC association in a comparison of TAC miR-133a-TG to sham nontransgenic hearts (Figure 6a–b, Supplemental Table 6c), suggesting that TAC competes with miR-133a overexpression for regulation of many mRNAs, in accordance with the second model, rather than behaving in an independent or additive manner. A similar proportion of the ‘tracking’ mRNAs (3 of the 32) in αMHC-miR-378 hearts maintained increased RISC associations during TAC (Figure 6e–f, Supplemental Table 7c) further demonstrating competition between TAC effects and microRNA overexpression.
Figure 6.
TAC effect on microRNA targets identified from RISC-sequencing in unstressed hearts. A–D) Venn analyses of direct (left panels) and indirect mRNA targets (right panels) of miR-133a in unstressed hearts (yellow circles), compared to mRNAs regulated in a RISC-dependent or RISC-independent manner in TAC αMHC-miR-133a hearts (red circles). E–H) As for A–D), but in αMHC-miR-378 hearts and with blue circles denoting miR-378 targets in unstressed hearts. mRNAs designated as ‘tracking’ mRNAs (not affected by TAC alone in panels A, C, E and G) are evaluated for similar regulation during TAC in the presence of the microRNA transgene (panels B, D, F and H).
As only a small proportion of microRNA transgene-regulated, RISC-associated mRNAs showed similar changes under sham and TAC conditions, we reasoned that most of the mRNAs exhibiting indirect, downstream regulation due to the microRNA transgenes would also be unlikely to undergo similar regulation under sham and TAC conditions. As for the set of 37 directly RISC-regulated 37 mRNAs used to track miR-133a targeting during TAC, we defined 190 mRNAs that were indirectly regulated (either upward or downward) by miR-133a under sham conditions without exhibiting similar regulation due to TAC alone. However, only 31 of these (16%) were similarly regulated by the combined stimuli of miR-133a overexpression and TAC (Figure 6c–d, Supplemental Table 6d). In similar fashion, 491 mRNAs were identified (at FDR<0.02) that were indirectly regulated by miR-378 overexpression but were not affected by TAC alone; 262 of these (53%) maintained similar regulation when TAC was performed on αMHC-miR-378 mice (Figure 6g–h, Supplemental Table 7d).
We did observe that while miR-133a and miR-378 precursors and their mature microRNA products continued to be overexpressed compared to wild-type levels during TAC, there was an apparent decline in transgene expression levels in RNA extracted from intact heart (Figure 5b). Firstly, as we noted when evaluating hypertrophic ‘marker’ RNAs during TAC (Figure 3c), pressure overload causes a significant decrease in whole-heart αMHC/Myh6 levels after 2 weeks and it is reasonable to expect that the transgene promoter would follow suit. Secondly, as referred to previously, FACS studies have demonstrated that nonmyocytes (especially fibroblasts) proliferate during the initial phase of TAC and contribute more substantially to total cardiac mass (and thus RNA) as a result42. Thus, it is likely that at least some, if not a substantial fraction, of the apparent decline in Myh6-directed microRNA transgene activity measured from whole-heart preparations during TAC is a result of increased nonmyocyte numbers and their contribution to total RNA.
Taken together, these data show that miR-133a and miR-378 regulation of direct, RISC-associated targets and of indirect, secondarily-affected mRNAs observed under unstressed conditions is largely overwhelmed by TAC-dependent transcriptional reprogramming, thus supporting the hypothesis that TAC-induced changes in mRNA abundances and mRNA RISC associations influence the biological function of microRNAs. Nonetheless, the above analyses did not investigate whether miR-133a or miR-378 overexpression could provoke changes in distinct subsets of RISC-associated and RISC-independent mRNAs in TAC that were not observed to be so affected under unstressed conditions. To evaluate this further possibility, and to seek to better understand the mechanisms underlying the previously observed salutary effects of miR-133a overexpression during TAC21, 22, we performed a pairwise comparison of RISC-associated and RISC-independent mRNA regulation during TAC in both αMHC-miR-133a mice and nontransgenic controls.
Distinct effects of miR-133a overexpression on pressure overload mRNA regulation
We recapitulated the previously reported decline in TUNEL-positive, apoptotic myocytes in αMHC-miR-133a hearts after 1 week of TAC and the lack of effect of the miR-133a transgene on the extent of hypertrophy21 (Supplemental Figure 6). Independent studies from another group using an inducible αMHC-miR-133a mouse line have also replicated the effect on fibrosis and implicated miR-133a suppression of members of the β1-adrenergic receptor signaling cascade from in vitro studies and biotin pulldown from heart homogenates22. However, our in vivo RISC-sequencing data under unstressed conditions did not uncover further evidence of direct miR-133a targeting of members of this cascade (Supplemental Table 6).
Numerous mRNA regulatory events observed during TAC were restricted to nontransgenic or to αMHC-miR-133a hearts. Only 38% (371 of 970) of direct microRNA targets, and 32% of RISC-independent targets (1202 of 3807) were shared by TAC in either genotype (Supplemental Figure 7a, 7b). Thus, the interaction of TAC and miR-133a overexpression is nonlinear: TAC is able to override effects of miR-133a overexpression observed in unstressed hearts, while the miR-133a transgene alters the transcriptome typically induced by TAC, restricting some regulatory events while synergizing with TAC to produce new ones. To focus on differences in apoptotic signaling between αMHC-miR-133a TAC and nontransgenic TAC, we performed further analyses of only those genes with Gene Ontology biological process annotations43 including the keyword ‘apoptosis’, using these as input to construct a knowledge-based signaling network44 (Supplemental Figure 8). Both nontransgenic and αMHC-miR-133a TAC gave rise to multiple interacting pro- and anti-apoptotic signals. While it was beyond the scope of these studies to elucidate which amongst these multiple altered signaling events most strongly favor cardiomyocyte survival during αMHC-miR-133a TAC, these data demonstrate differential regulation of multiple apoptotic effectors and protectors in response to elevated miR-133a. In combination with the observed phenotype of αMHC-miR-133a TAC hearts, these data suggest that the decrease in miR-133a observed during TAC in wild-type animals may rebalance these multiple pathways such that cell death is favored.
Distinct effects of miR-378 overexpression on pressure overload mRNA regulation
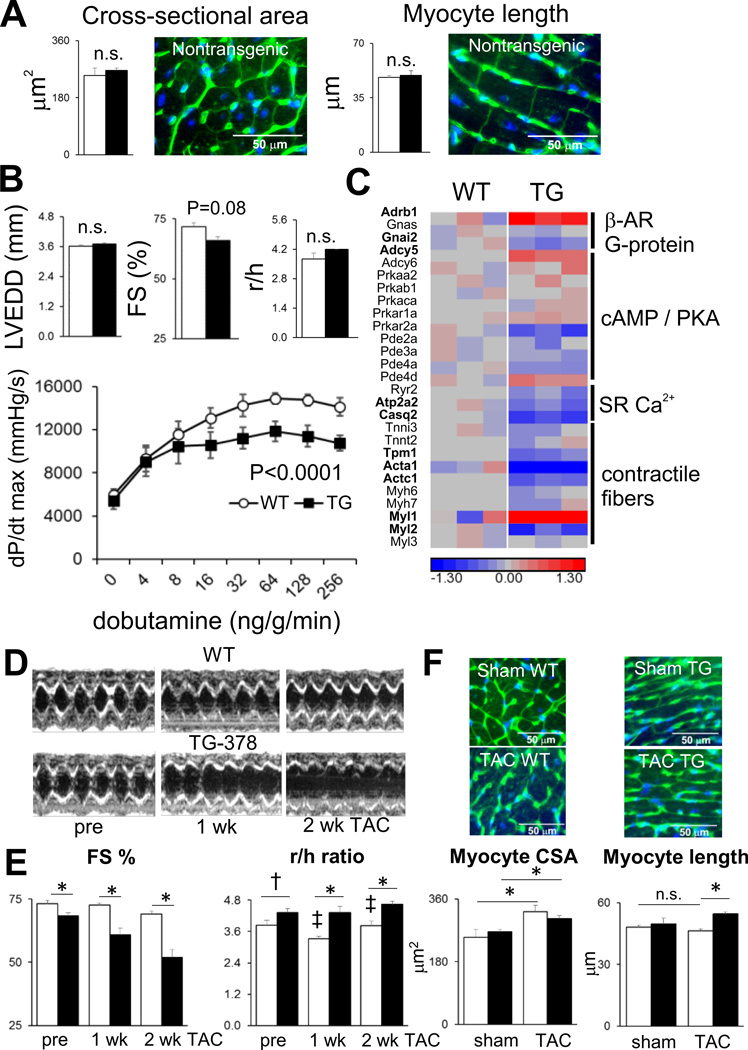
In similar fashion to αMHC-miR-133a hearts, numerous mRNA regulatory events observed during TAC were restricted to nontransgenic or to αMHC-miR-378 hearts. Only 5% (7 of 146) of direct microRNA targets, and 15% of RISC-independent targets (220 of 1427) were shared by TAC in either genotype (Supplemental Figure 7c, 7d). However, in contrast to the miR-133a studies, the phenotype of αMHC-miR-378 mice (other than the effect of the transgene on the production of other microRNAs27) has not been previously described, either in unstressed or stressed conditions. Prior to these experiments, we had hypothesized that genetically increased levels of cardiomyocyte miR-378, in the context of pressure overload, may serve to limit myocyte hypertrophy, myocyte death, and replacement fibrosis, similar to AAV9-mediated transduction of miR-378 to the heart24. There were no changes in myocyte cross-sectional area or length, or in cardiac chamber volume, in unstressed αMHC-miR-378 hearts (Figure 7a). Slight decreases in echocardiographic fractional shortening, and in dobutamine-stimulated contractility measured during cardiac catheterization, were evident (Figure 7b). At the global RNA abundance level, the molecular basis of the reduced contractility did not appear to involve downregulation of Adrb1 (β1-adrenergic receptor), G proteins or adenylyl cyclase enzyme mRNAs but may depend, at least in part, on decreases in other key components of the contractile machinery including calsequestrin 2 (Casq2), tropomyosin I (Tpm1), cardiac alpha-actin (Actc1) and cardiac myosin light chain 2 (Myl2) (Figure 7c).
Figure 7.
Effect of miR-378 overexpression on cardiac contractile performance and TAC response. A) Myocyte cross-sectional area (from ~500 cells/heart) and length (from ~100 cells/heart); representative images from nontransgenic hearts are shown. B) M-mode echocardiography in awake mice (n=6–10 per group) and dobutamine-stimulated contractility in anesthetized mice, n=4 mice per group; P<0.0001 between genotypes by 2-way ANOVA with Sidak multiple correction test. C) β-adrenergic pathway and contractile function mRNAs; heatmap colors show log2fold changes relative to nontransgenic mean. Significantly regulated mRNAs (fold-change 25%, FDR<0.1) are in bold. D) Representative M-mode echocardiograms and E) echocardiographic parameters determined from TAC studies. F) Myocyte cross-sectional area and lengths as for A). White bars, nontransgenic; black bars, αMHC-miR-378. * = P<0.05, † = P<0.1, unpaired 2-tailed t-test; ‡ = P<0.05 relative to previous time point; n=7–8 per group.
In contrast to the outcomes of AAV9-miR-378 and anti-miR-378 studies24, 39, αMHC-miR-378 hearts exhibited a rapid decline in functional shortening and did not appear to undergo initial wall thickening (decreased r/h ratio) in response to pressure overload, unlike their wild-type controls (Figure 7d, 7f). Similar increases were observed in myocyte cross-sectional area, but myocyte length was increased only in transgenic mice subjected to pressure overload (Figure 7e). An increased r/h ratio measured via echocardiography was observed in parallel to the increase in sectioned myocyte length (Figure 7f). The extent to which further molecular, mechanistic dissection of the TAC phenotype of αMHC-miR-378 mice may be worthwhile is mitigated by the fact that αMHC-miR-378 hearts exhibit ~12-fold miR-378 overexpression during TAC (Figure 5), in contrast to AAV9-miR-378-treated hearts in which miR-378 expression during TAC is essentially normalized in comparison to pre-TAC conditions24. Nonetheless, superposition of TAC in two different strains of microRNA-overexpressing hearts demonstrated that TAC altered microRNA-mRNA targeting relationships, regardless of whether the combined effect of TAC and microRNA overexpression was modest (miR-133a) or deleterious (miR-378).
Discussion
The primary goal of these studies was to determine whether mRNA-RISC associations mediated by individual microRNAs are the same under two different physiological conditions; a basal, unstressed state and pressure overload hypertrophy. While in silico or in vitro protocols can partially estimate the likelihood that a given mRNA-RISC binding event is able to occur, they typically do not easily allow the stoichiometries of mRNAs and of microRNAs/RISCs to be factored in. Consequentially, it is difficult to estimate the proportion of an individual microRNA that might be bound to each of several potential mRNA targets, and which of several microRNAs might constitute the most potent regulator of an individual mRNA; factors which are likely to be highly sensitive to cell type and status. Here, we have employed RISC-sequencing of intact hearts to monitor changes in mRNA targeting by Ago2 during cardiac stress, in combination with microRNA-expressing transgenes to be able to accurately track microRNA-bound mRNAs. Overexpression of miR-133a and miR-378 provided a means of tracking microRNA-targeted mRNAs in unstressed and stressed hearts so that the primary goal of these studies could be achieved. Our findings demonstrate that some microRNA-targeted mRNAs in unstressed conditions no longer undergo RISC-mediated suppression during TAC, while other mRNAs not subject to RISC-mediated suppression in unstressed conditions become so during TAC. Thus, the signals influenced by particular microRNAs depend on precise tissue status, including the manner in which the remainder of the transcriptome is expressed, a parameter which will vary in health or disease. In consequence, we propose that rational design of microRNA-based interventions in cardiac disease needs to be based not principally on mRNA suppression events observed in vitro or in unstressed hearts, but needs to take into account whether the same events continue to occur in diseased hearts and whether inhibiting events that only occur in diseased hearts may provide new opportunities.
The use of microRNA overexpression combined with RISC-sequencing in vivo may identify a greater range of cardiac targets of miR-133a and miR-378 in health or disease than is readily possible with in vitro or in silico approaches. Nonetheless, as we have previously discussed, RISC-sequencing of microRNA transgenic hearts may not identify those targets of a microRNA that are already comprehensively suppressed in wild-type hearts16, 17, and microRNA transgenes that provoke disease alone or exacerbate disease responses severely complicate the analysis of endogenous microRNA targets in these contexts17. In addition, the use of transgenes that encode microRNA stem-loop precursors means that both the more abundant ‘guide’ microRNA strand and the less abundant ‘passenger’ microRNA strand are overexpressed, each of which could have an independent set of mRNA targets27. We have shown previously that a range of other microRNA species are dysregulated in αMHC-miR-378 hearts and could be responsible for some of the effects observed on the mRNA transcriptome27. Thus, the direct applicability of our transgenic miR-133a and miR-378 targeting profiles for accurate definition of the mRNAs targeted by miR-133a and miR-378 in wild-type hearts is limited by how faithfully these targeting profiles reflect the wild-type context. More accurate determination of authentic miR-133a and miR-378 direct and indirect mRNA targets in healthy and stressed hearts, using mRNA-RISC interaction studies in vivo, may need to involve comparative studies in hearts lacking some, but not all miR-133a alleles45, in hearts lacking miR-37846, and/or in hearts with temporal control of microRNA transgenes22 or shorter-term microRNA manipulations made possible by viri20, 24 or anti-miRs11, 20, 39.
It could be argued that the fastest path to achieving useful therapies based on microRNA actions47 might be to work primarily to reverse disease-mediated microRNA changes with the expectation that the complex signals they regulate will regain equilibrium (e.g. reversal of dysregulated miR-34a9, miR-199b11, miR-212/13213, and miR-37824 in hypertrophic hearts). Indeed, several such preclinical studies have already reported success in ameliorating consequences of pressure overload. Nonetheless, a more refined approach of altering particular microRNA-mRNA interactions (e.g. with ‘decoy’ target-protection oligonucleotides designed to disrupt specific binding events48) made possible by detailed studies of the kind outlined here, may prove to be highly valuable as an alternative to affecting the entire targeting portfolio of a given microRNA. This presents a need for more specific determination of microRNA binding sites and suppressive actions on mRNAs in vivo. Crosslinking prior to Ago2 immunoprecipitation, in combination with rigorous analysis of crosslinking-induced sequence mutations, can provide a comprehensive view of the binding of individual microRNAs to individual mRNAs49, but has not yet been demonstrated using intact hearts, nor has its sensitivity in differential expression studies been evaluated. Nonetheless, this or related approaches capable of interrogating endogenous in vivo interactions would avoid many of the complications arising from genetic or reagent-based microRNA overexpression and knockout. Continued methodological development and training of informatic helper algorithms with contextually appropriate empirical data should lead to a greater understanding of the actions of individual microRNAs, whether stress-regulated or of stable abundance, in health and disease. In turn, this will lead to optimized rationales for choosing individual microRNAs for therapeutic manipulation.
Supplementary Material
Clinical Perspective.
While microRNAs are known to possess important functions in cardiac stress responses, the mRNAs that are primarily affected by changes in cardiac microRNAs are not well defined. Since RNA binding relationships depend on compatible nucleotide sequences as well as complex stoichiometries, stress-dependent large-scale alterations in the cardiac transcriptome during disease suggest that the mRNAs most substantively targeted by individual microRNAs will vary between unstressed and stressed conditions. To test the hypothesis that microRNA target profiles differ in health and disease, we traced the fate of empirically-determined miR-133a and miR-378 targets in mouse hearts undergoing pressure-overload hypertrophy, using next-generation RNA-sequencing techniques to define cohorts of mRNAs regulated in a microRNA-dependent or –independent manner. We found that only a small proportion of mRNAs targeted by either miR-133a or miR-378 in unstressed conditions continued to be targeted by these same microRNAs in pressure-overload hypertrophy. Thus, pressure-overload alters individual microRNA targeting profiles, concomitant with widespread changes in other cardiac RNAs. This reinforces the need to define microRNA targets in tissue-, cell- and status-specific contexts for a proper understanding of microRNA-dependent contributions to signaling pathways. Furthermore, it suggests that planned microRNA-based interventions in pathological circumstances may need to be based primarily on RNA interaction data obtained from hearts exhibiting similar pathology, to permit an optimal choice of individual microRNAs for therapeutic manipulation.
Acknowledgments
Funding Sources
Research reported in this publication was supported by American Heart Association Grant-in-Aid 14GRNT20410000 and Washington University Department of Medicine funds to S.J.M., NIH R01 HL108943 to G.W.D., and by NIH UL1 TR000448 to the Washington University Institute of Clinical and Translational Sciences from the National Center for Advancing Translational Sciences (NCATS).
Footnotes
Conflict of Interest Disclosure: None
References
- 1.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 2.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 3.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 5.Khorshid M, Hausser J, Zavolan M, van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods. 2013;10:253–255. doi: 10.1038/nmeth.2341. [DOI] [PubMed] [Google Scholar]
- 6.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 7.Karakikes I, Chaanine AH, Kang S, Mukete BN, Jeong D, Zhang S, et al. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J Am Heart Assoc. 2013;2:e000078. doi: 10.1161/JAHA.113.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayed D, Yang Z, He M, Pfleger JM, Abdellatif M. Acute targeting of general transcription factor IIB restricts cardiac hypertrophy via selective inhibition of gene transcription. Circ Heart Fail. 2015;8:138–148. doi: 10.1161/CIRCHEARTFAILURE.114.001660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seok HY, Chen J, Kataoka M, Huang ZP, Ding J, Yan J, et al. Loss of MicroRNA-155 protects the heart from pathological cardiac hypertrophy. Circ Res. 2014;114:1585–1595. doi: 10.1161/CIRCRESAHA.114.303784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol. 2010;12:1220–1227. doi: 10.1038/ncb2126. [DOI] [PubMed] [Google Scholar]
- 12.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Matkovich SJ, Hecker PA, Zhang Y, Dorn GW., II Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac microRNAs. Proc Natl Acad Sci USA. 2012;109:19864–19869. doi: 10.1073/pnas.1214996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdellatif M. Differential expression of microRNAs in different disease states. Circ Res. 2012;110:638–650. doi: 10.1161/CIRCRESAHA.111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matkovich SJ, Hu Y, Eschenbacher WH, Dorn LE, Dorn GW., II Direct and indirect involvement of microRNA-499 in clinical and experimental cardiomyopathy. Circ Res. 2012;111:521–531. doi: 10.1161/CIRCRESAHA.112.265736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorn GW, 2nd, Matkovich SJ, Eschenbacher WH, Zhang Y. A human 3' miR-499 mutation alters cardiac mRNA targeting and function. Circ Res. 2012;110:958–967. doi: 10.1161/CIRCRESAHA.111.260752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 21.Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, et al. MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaldi A, Zaglia T, Di Mauro V, Carullo P, Viggiani G, Borile G, et al. MicroRNA-133 modulates the beta1-adrenergic receptor transduction cascade. Circ Res. 2014;115:273–283. doi: 10.1161/CIRCRESAHA.115.303252. [DOI] [PubMed] [Google Scholar]
- 23.Nagalingam RS, Sundaresan NR, Gupta MP, Geenen DL, Solaro RJ, Gupta M. A cardiac-enriched microRNA, miR-378, blocks cardiac hypertrophy by targeting Ras signaling. J Biol Chem. 2013;288:11216–11232. doi: 10.1074/jbc.M112.442384. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation. 2013;127:2097–2106. doi: 10.1161/CIRCULATIONAHA.112.000882. [DOI] [PubMed] [Google Scholar]
- 25.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matkovich SJ, Hu Y, Dorn GW., 2nd Regulation of cardiac microRNAs by cardiac microRNAs. Circ Res. 2013;113:62–71. doi: 10.1161/CIRCRESAHA.113.300975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 30.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 31.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, et al. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Huang W, Xu R, Nie Y, Cao X, Meng J, et al. MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J Cell Mol Med. 2012;16:2150–2160. doi: 10.1111/j.1582-4934.2012.01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li RC, Tao J, Guo YB, Wu HD, Liu RF, Bai Y, et al. In vivo suppression of microRNA-24 prevents the transition toward decompensated hypertrophy in aortic-constricted mice. Circ Res. 2013;112:601–605. doi: 10.1161/CIRCRESAHA.112.300806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahlquist C, Jeong D, Rojas-Munoz A, Kho C, Lee A, Mitsuyama S, et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508:531–535. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 36.Pan Z, Sun X, Shan H, Wang N, Wang J, Ren J, et al. MicroRNA-101 inhibited postinfarct cardiac fibrosis and improved left ventricular compliance via the FBJ osteosarcoma oncogene/transforming growth factor-beta1 pathway. Circulation. 2012;126:840–850. doi: 10.1161/CIRCULATIONAHA.112.094524. [DOI] [PubMed] [Google Scholar]
- 37.Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagalingam RS, Sundaresan NR, Noor M, Gupta MP, Solaro RJ, Gupta M. Deficiency of cardiomyocyte-specific-microRNA-378 contributes to the development of cardiac fibrosis involving a TGF b1-dependent paracrine mechanism. J Biol Chem. 2014;289:27199–27214. doi: 10.1074/jbc.M114.580977. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW., 2nd Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C, Dorn GW., 2nd Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci USA. 2014;111:12264–12269. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Souders CA, Borg TK, Banerjee I, Baudino TA. Pressure overload induces early morphological changes in the heart. Am J Pathol. 2012;181:1226–1235. doi: 10.1016/j.ajpath.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Reuters Thomson. MetaCore. [Accessed 4/2/14]; Available at: http://www.genego.com. [Google Scholar]
- 45.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci USA. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 48.Drawnel FM, Wachten D, Molkentin JD, Maillet M, Aronsen JM, Swift F, et al. Mutual antagonism between IP(3)RII and miRNA-133a regulates calcium signals and cardiac hypertrophy. J Cell Biol. 2012;199:783–798. doi: 10.1083/jcb.201111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9:263–293. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.