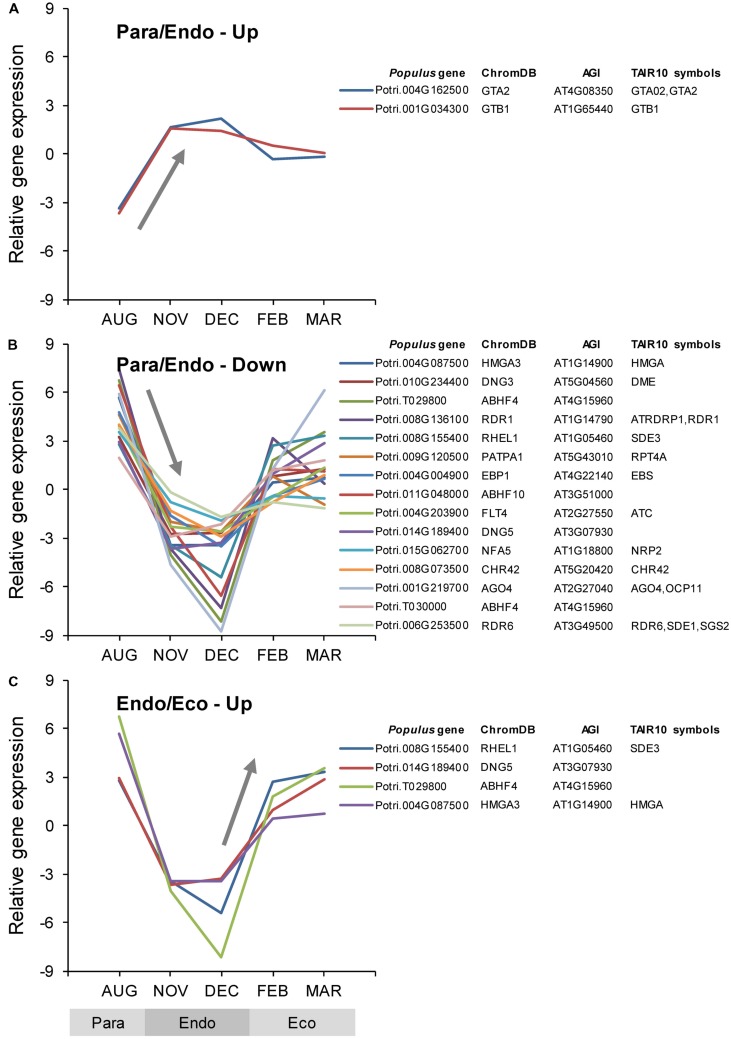
FIGURE 3.
Relative expression of differentially expressed chromatin-associated genes. Chromatin-associated genes were identified using the Arabidopsis thaliana chromatin database (ChromDB; Gendler et al., 2008). Genes were classified into four groups: up-regulated or down-regulated from paradormancy to endodormancy (A,B), or up-regulated or down-regulated from endodormancy to ecodormancy (C). Within each group, we ranked genes by FDR p-value, and then displayed the top 15 genes for each group if they had a FDR p-value < 0.05. Only four genes were differentially expressed between endodormancy and ecodormancy, and all of these were up-regulated (C). Gene expression values are the means for each month normalized to a mean of zero and a standard deviation equal to the ANOVA RMSE from the analysis of gene expression differences among months. ChromDB is the ChromDB identifier, Populus gene is the P. trichocarpa gene-locus name, and AGI and TAIR10 symbols are the Arabidopsis gene identifiers and gene symbols from the P. trichocarpa v3.0 annotations (http://www.phytozome.net/).