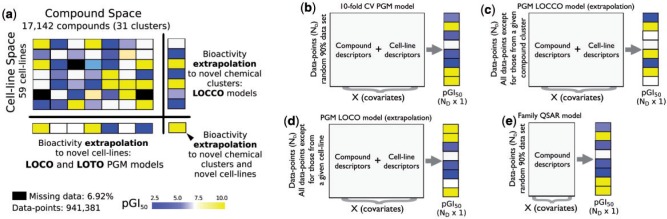
Fig. 1.
Pharmacogenomic modelling concept and illustration of the learning strategies explored. (a) The pGI50 values for 17 142 compounds on 59 cancer cell lines (941 831 data points) were modelled with RF and SVM models and conformal prediction. (b) Illustration of the training data used in the following learning strategies: (b) 10-fold CV PGM models (interpolation); (c) LOCCO; (d) LOCO; and (e) Family QSAR. As can be seen in (b–e), the training data used in each learning strategy differs with respect to (i) the subset of data-points from the whole dataset used for training and (ii) the type and combination of input descriptors, which can be only compound descriptors, only cell line descriptors, or the combination of both. In all models reported in this article, Morgan fingerprints were used as compound descriptors, whereas the dataset views indicated in Table 1 and four cell line kernels were used to encode the cell lines. Overall, this validation enabled us to assess the model’s performance in real-world settings, where the extrapolation to novel cell lines and compounds is often a necessary step