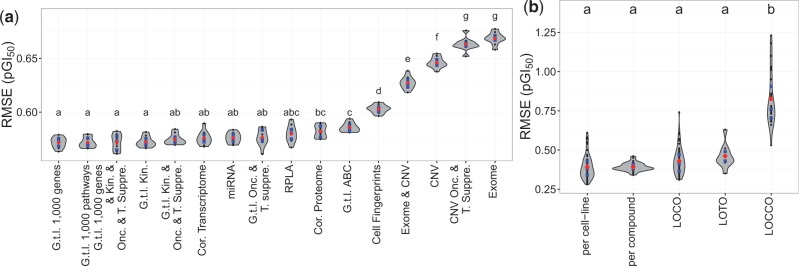
Fig. 3.
Benchmarking the cell line profiling dataset views for the cell line sensitivity prediction. (a) The predictive power of the 14 dataset views (Table 1) and two cell line kernels, namely cor. Proteome and cor. Transcriptome, was quantified by the RMSE values on the test set. For each dataset view, we trained the 10-fold CV PGM models on the uncorrelated bioactivities 0.5 dataset. We found significant differences among the dataset views (ANOVA, P < 0.01). Post-hoc analyses (Tukey’s HSD, α = 0.05) were used to cluster the dataset views according to their predictive power. Dataset views sharing a letter label performed at the same level of statistical significance. We consistently found that the gene transcript levels and the abundance of proteins and miRNA led to the most predictive models (labelled with ‘a’). (b) The evaluation of both interpolation and extrapolation power was evaluated on the complete dataset. After finding significant differences across groups (ANOVA, P < 0.01), we found that the PGM models interpolate and extrapolate to new cell lines and tissues at the same level of statistical significance (Tukey’s HSD, α = 0.05). In contrast, we found statistically significant differences in the performance between extrapolation and interpolation to new chemical clusters. The blue points indicate the median and the interquartile range (25th–75th percentile), whereas the red points indicate the mean RMSE value