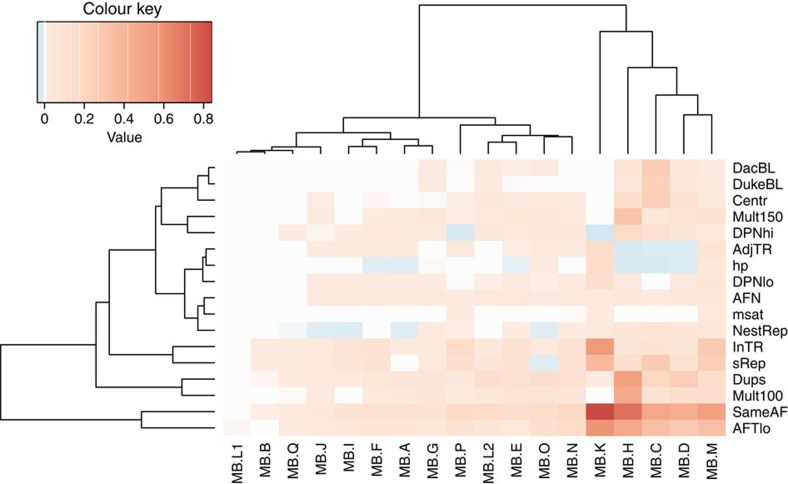
Figure 6. Enrichment or depletion of genomic and alignment features in FP calls for each medulloblastoma SSM submission.
For each feature, the difference in frequency with respect to the Gold Set is multiplied by the FP rate. Blue indicates values less than zero and thus the proportion of variants or their score on that feature is lower in the FP set with respect to the true variants. Reddish colours correspond to a higher proportion of variants or higher scores for the feature in FP calls versus the Gold Set. Both features and submissions are clustered hierarchically. The features shown here include same AF (the probability that the AF in the tumour sample is not higher than that in the normal samples, derived from the snape-cmp-counts score), DacBL (in ENCODE DAC mappability blacklist region), DukeBL (in Encode Duke Mappability blacklist region), centr (in centromere or centromeric repeat), mult100 (1—mappability of 100mers with 1% mismatch), map150 (1—mappability of 150mers with 1% mismatch), DPNhi (high depth in normal), DPNlo (low depth in normal), dups (in high-identity segmental duplication), nestRep (in nested repeat), sRep (in simple repeat), inTR (in tandem repeat), adjTR (immediately adjacent to tandem repeat), msat (in microsatellite), hp (in or next to homopolymer of length >6), AFN (mutant AF in normal) and AFTlo (mutant AF in tumour<10%).