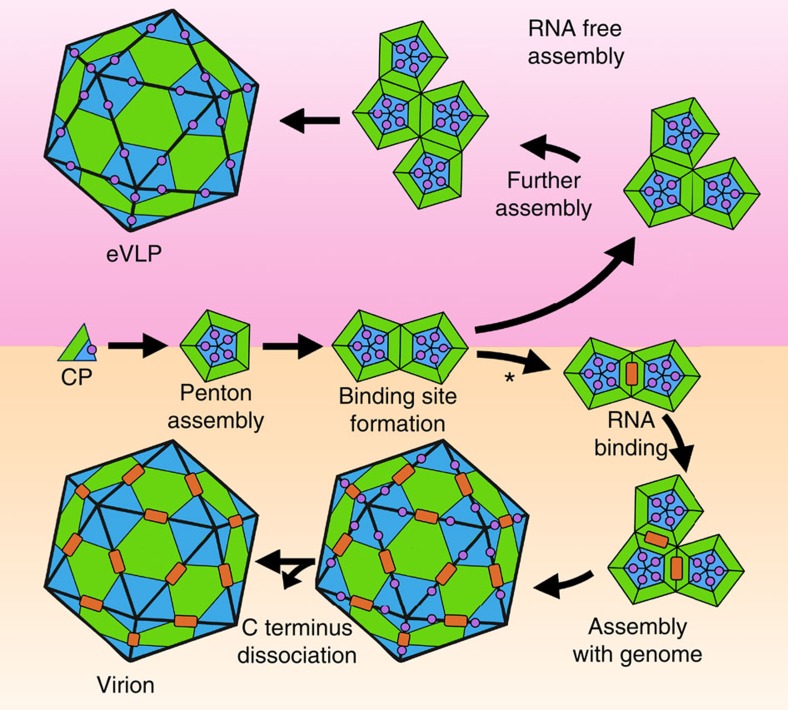
Figure 8. A model for CPMV Assembly. The L (green) and S (blue) subunits associate to form a coat protein penton.
The C-terminal extension (magenta circle) appears to act as a dab of ‘molecular glue', stabilizing the formation of the penton due to extensive interactions with the neighbouring S subunit. In the absence of RNA (RNA free assembly shown in the top half of the diagram), C-terminal cleavage is slow, and eVLP particles are produced. After time, the C-terminal extension is lost from the eVLP, suggesting that once an extensive protein:protein interaction network is formed in the capsid the eVLP structure is stable. In the presence of genomic RNA (assembly with genome shown in the bottom half of the diagram), assembly follows a different path. Initially, pentons are formed in the same way as in the eVLP, using the C-terminal extension for stability. Two pentons interact to form a twofold axis, and thus the RNA-binding site appears to exist at that position. Our model shows genomic RNA (represented as an orange rectangle) initially binds to the two pentons at this twofold axis, the first step in genome encapsidation. Following stepwise addition of pentons, the C terminus is rapidly cleaved implying that RNA occupancy is allosterically communicated through the structure to the outside of the capsid.