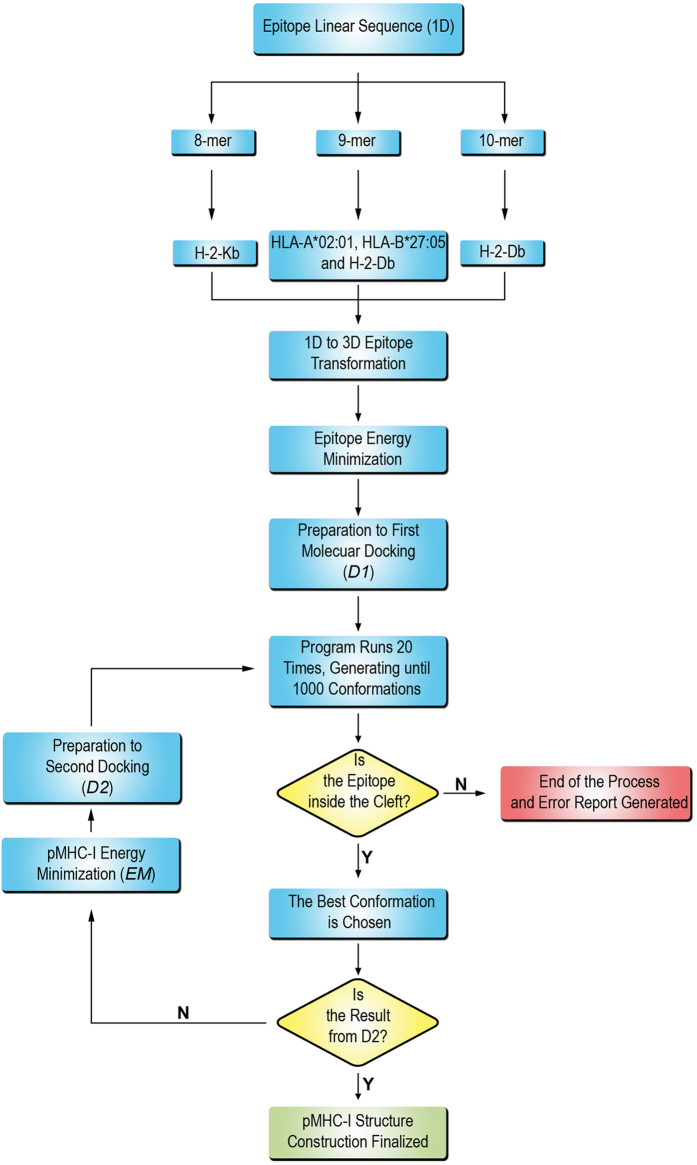
Figure 1. Flowchart showing the DockTope sequence of steps.
The program starts from the linear sequence of the epitope and terminates when the pMHC-I structure is obtained. The user chooses among the allotypes HLA-A*02:01, HLA-B*27:05, H-2-Db and H-2-Kb, which depends on the epitope length (8-mer, 9-mer or 10-mer). The program prepares the files for the first docking (D1), where the best suited conformations will be saved during 20 rounds of simulation. The program checks whether the epitope is inside the cleft. In case of error, a report is written and the program stops. Otherwise, the program proceeds to the next step, where the best conformation is chosen. The program checks whether the structure generated is coming from D1. If so, the structure is energy minimized (EM) and a second docking is performed. In the end, the pMHC-I structure is generated in the PDB format.