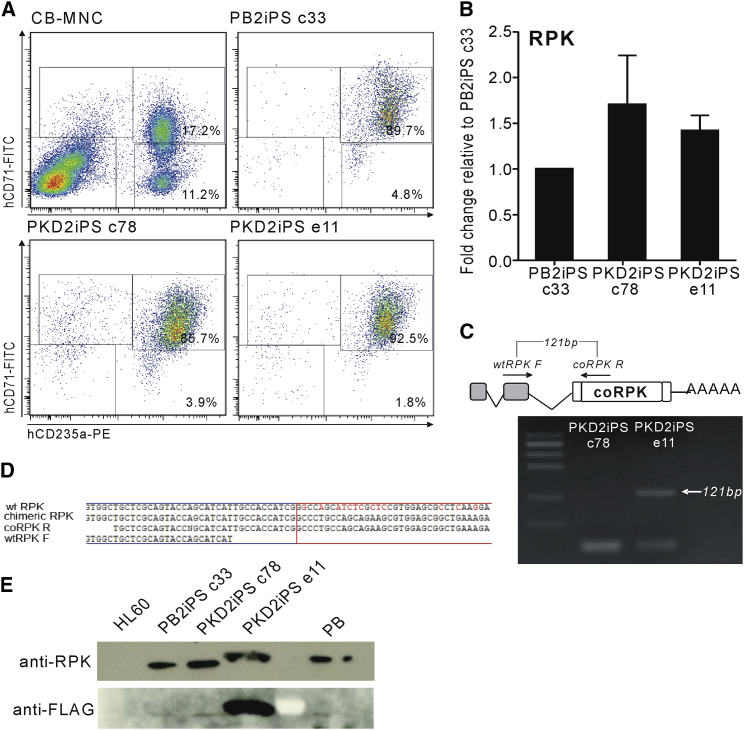
Figure 4.
Erythroid Differentiation of PKD2iPSCs
PB2iPSCs, PKD2iPSCs, and edited PKD2iPSCs were differentiated to erythroid cells under specific conditions and analyzed after 31 days in in vitro proliferation and differentiation conditions.
(A) Erythroid differentiation was confirmed by flow cytometry analysis. Cord blood MNCs, PB2iPSC clone c33, PKD2iPC clone c78, and edited PKD2iPSC clone e11 representative analyses are shown.
(B) RPK expression in erythroid cells derived from the different iPSCs was evaluated by qRT-PCR (n = 6).
(C) Specific RT-PCR to amplify the chimeric (mRNA)RPK in edited PKD2iPSC. The primers amplified the region around the link between endogenous (mRNA)RPK and the introduced codon-optimized (cDNA)RPK sequence. Arrow indicates the expected band and the corresponding size only preset in the RNA from edited cells (PKD2iPSC e11).
(D) The sequence of the chimeric transcript was aligned with the theoretical expected sequence after the correct splicing between the endogenous exon 2 (blue square) and the exogenous exon 3 (red square).
(E) The presences of RPK protein in erythroid cells derived from PB2iPSCs, PKD2iPSCs, and edited PKD2iPSCs assessed by western blot (upper line); mobility change in PKD2iPSC e11 is due to the FLAG tag added to the chimeric protein. Expression of chimeric protein was detected by anti-FLAG antibody only in erythroid cells derived from edited PKD2iPSCs (bottom line).
See also Figures S5 and S6.