Figure 6.
Transcriptome Analysis of Different Subsets of Tonsillar Epithelial Cells
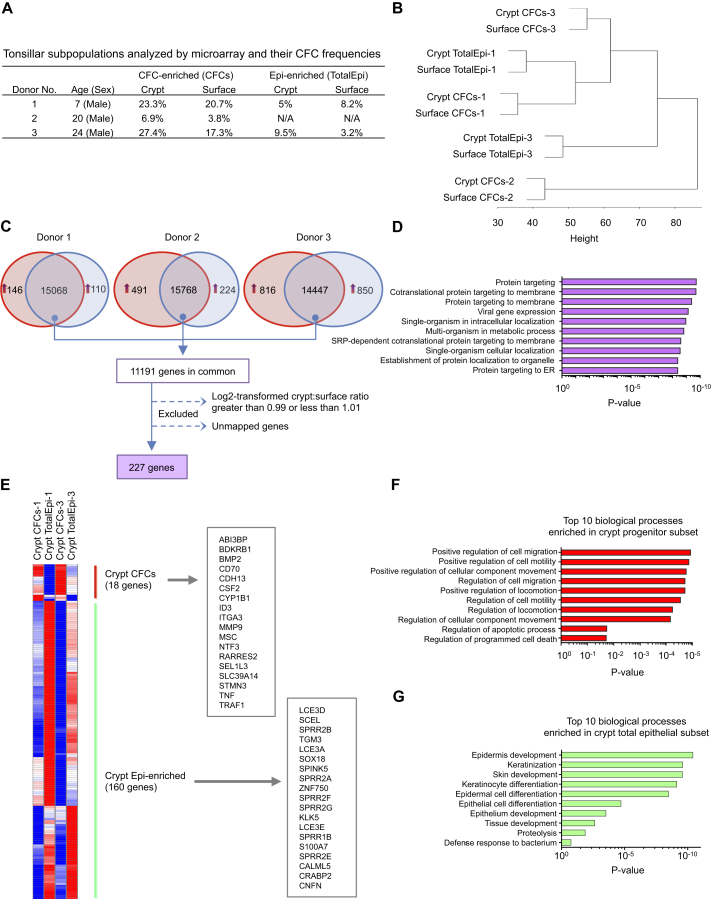
(A) The age and sex of the donors and colony-forming efficiencies of the freshly isolated CD44+NGFR+ cells (CFCs) and CD45−CD31− total epithelial cells (TotalEpi) used for the transcriptome analyses.
(B) Illumina transcriptome analyses identifying the relatedness of all four subsets of tonsillar cells derived from an unsupervised hierarchical clustering analysis.
(C) Number of genes expressed in crypt CD44+NGFR+ cells and donor-matched surface TotalEpi subsets obtained from three donors. The number of genes more highly expressed in crypt or in surface by at least a 1.5-fold differential is shown in the red and blue areas, respectively. The number in the overlapping region represents the number of transcripts that displayed a <1.5-fold differential expression between the two sites. Those with crypt-to-surface ratios of log2-transformed values ≥0.99 and ≤1.01 were considered to be equally expressed and used for analysis.
(D) The top ten biological processes associated with genes equally expressed in surface and crypt CD44+NGFR+ subsets based on gene ontology assignment. SRP, signal recognition particle.
(E) Hierarchical cluster analysis of CD44+NGFR+ cells and their respective TotalEpi subsets generated by identifying the genes that displayed a 2-fold or higher differential expression in the Illumina data comparisons. Shown in red are genes that were highly overexpressed in CD44+NGFR+ cells in comparison with the TotalEpi populations. Shown in green are those more highly expressed in the two TotalEpi populations included in this comparison (n = 2, p ≤ 0.05).
(F) The top ten biological GO categories found to be upregulated in crypt CD44+NGFR+ cells compared with the TotalEpi populations.
(G) The top ten biological GO categories upregulated in the TotalEpi populations compared with CD44+NGFR+ crypt cells.
See also Figure S2.