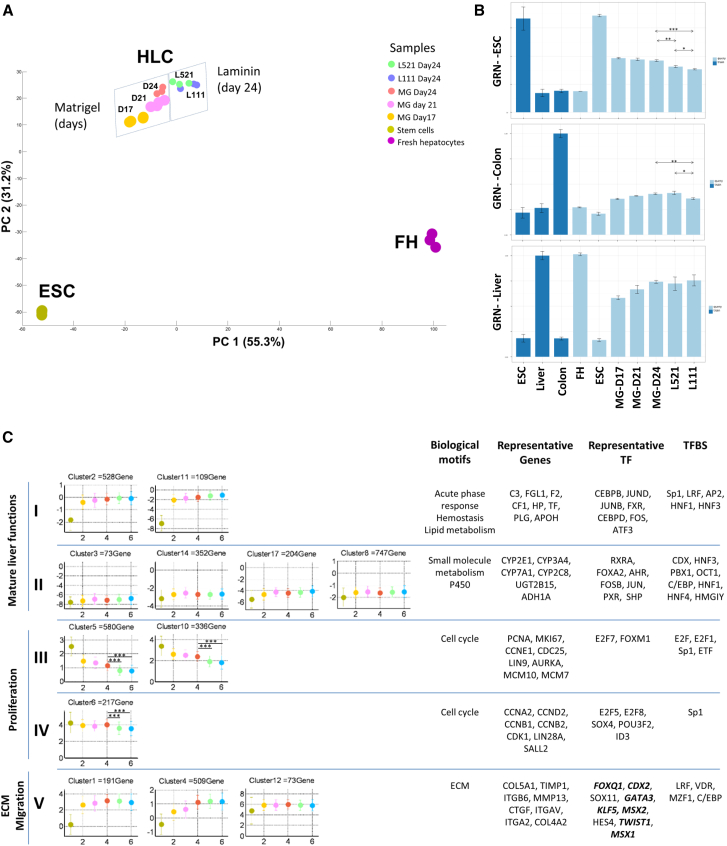
Figure 7.
Gene Array Analysis Identifies Genome-Wide Effects of the Laminin Matrix in HLC Differentiation
(A) Principal component analysis of the 1,000 genes with highest variance in hESCs, FHs, and HLCs differentiated on MG for 17, 21, and 24 days or L111 or L521 for 24 days. The results shown represent three biological replicates. The graph shows the two largest principal components that constitute 86.5% of the variance.
(B) GRN status obtained from the gene expression profile in FHs, ESCs and HLCs on the different matrices. The training score for ESCs, colon, and liver are shown in dark blue and represent the maximal score for each cell/tissue. The score for the queried samples (in light blue) is calculated in relation to the maximal cell/tissue specific score, as described in Cahan et al. (2014). Significantly different scores were observed for laminin 521 and laminin 111 versus Matrigel (day 24) on ESCs and Colon GRNs (∗∗∗p < 0.005, ∗∗p < 0.05, ∗0.5, t test).
(C) Clustering of genes with similar expression patterns in HLCs generated three superclusters representing the motifs “mature liver functions,” “proliferation,” and “ECM/migration” (see Supplemental Information for details). Shown are the KEGG and gene ontology (GO) motifs overrepresented in each gene cluster. Representative genes for each cluster and motif are also indicated. Genes in cluster V were expressed at lower levels on HLCs in laminin 111 compared with Matrigel. See Table S3 for a full list of gene and motif enrichment analyses in each gene cluster. Data are presented as the mean of three independent experiments. significant differences in mean gene expression levels (log2 scale) were observed between Matrigel (day 24), laminin 521, and laminin 111 in clusters 5, 10, and 6, which correspond to the supercluster proliferation (t test, p < 0.005).