Figure 1.
Selection of Optimal Ventricular Cardiomyocyte-Specific IRX4 Molecular Beacons
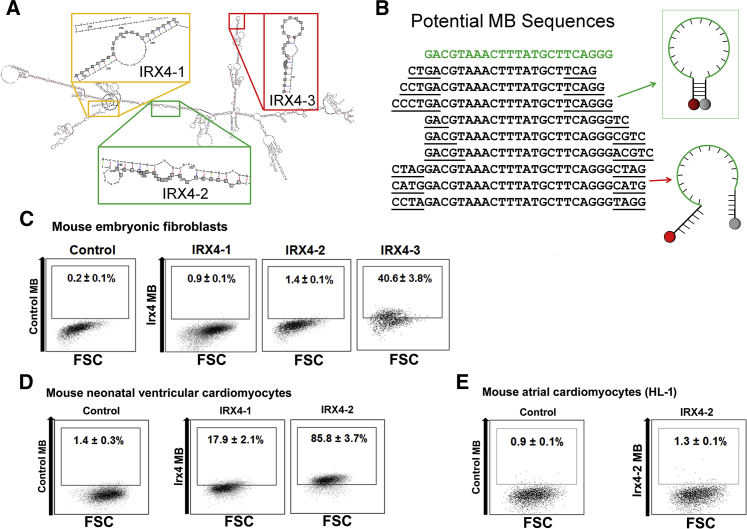
(A) Irx4 mRNA structure was predicted using the RNAfold web server. Three unique target sequences were identified in Irx4 mRNA that maximized the number of predicted unpaired bases as well as the binding affinity of a complementary probe.
(B) Stem sequences were appended to the complementary sequence and evaluated using QUIKFOLD to minimize the free energy that causes the oligonucleotide to assume a hairpin structure in solution.
(C–E) Flow cytometry results after delivering various IRX4 MBs designed to identify Irx4 mRNAs, or control MB, into mouse embryonic fibroblasts (C), eonatal mouse ventricular CMs (D), and HL-1 CMs (E). The number in each panel represents the percentage of fluorescent cells. FSC indicates forward scatter. All experiments were performed on three independent biological replicates (C–E).