Figure 1.
Phylogenetic Properties of MICU in Plants, Gene Model in Arabidopsis, and Ca2+ Binding.
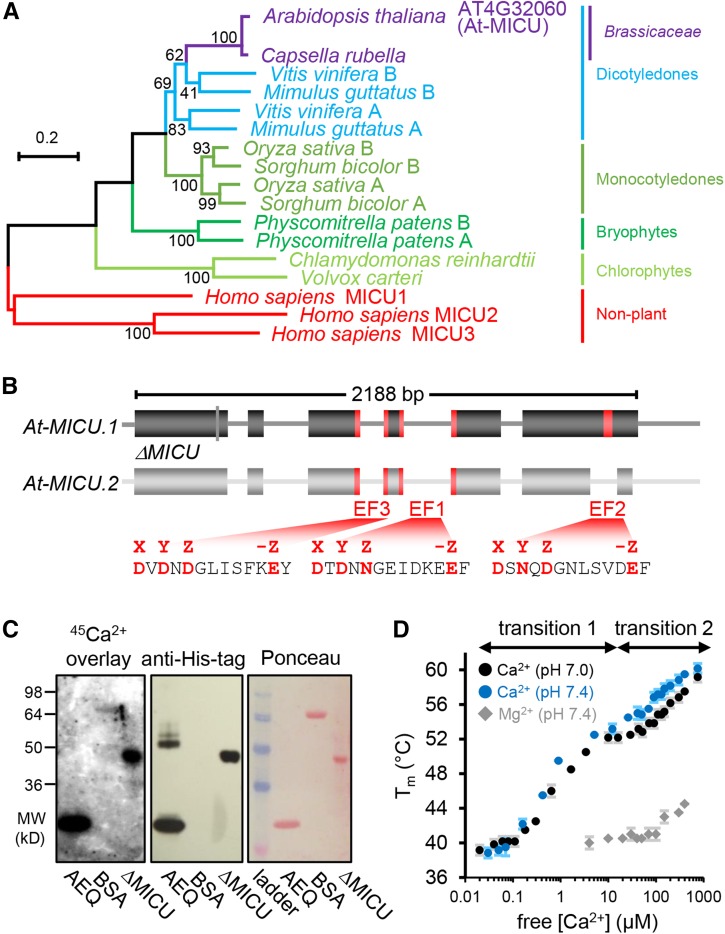
(A) Phylogenetic tree of homologs from representative plant species to human MICU proteins. The scale bar represents 20 substitutions per 100 amino acids. Bootstrap values (on nodes) were calculated using 1000 replicates. The alignments are shown in Supplemental File 1.
(B) Gene model of Arabidopsis MICU variants. MICU.1 and MICU.2 represent predicted splice forms; ΔMICU, a variant of MICU.1 cDNA where nucleotides coding for 117 N-terminal amino acids were removed to use in recombinant protein expression. Exons and introns are illustrated as boxes and lines, respectively. Nucleotides encoding predicted Ca2+ binding EF-hand motifs are highlighted in red. All three EF-hands are conserved among plants. EF1 and EF2 are also conserved in mammalian MICUs, while EF3 is not (Supplemental Figure 1). The amino acid sequences that include all critical charged residues (red) predict genuine Ca2+ binding capability for all three EF-hands.
(C) 45Ca2+ overlay assays of recombinantly purified ΔMICU protein (Supplemental Figure 2) compared with recombinant aequorin (AEQ) and BSA (BSA) as positive and negative control, respectively. The presence of protein was validated by immunodetection of the His-tags of ΔMICU and AEQ, and Ponceau S staining of separate control membranes. 45Ca2+ binding was visualized by autoradiography.
(D) Thermostabilization of ΔMICU through Ca2+. Protein melting points (Tm) were recorded in the presence of Ca2+/EGTA-adjusted free Ca2+ at pH 7.0 (black circles) and 7.4 (blue circles), and Mg2+/EDTA-adjusted free Mg2+ concentrations at pH 7.4 (gray diamonds). Concentration ranges of free Ca2+ associated with clear Tm transition are indicated. n = 3 (2 for Mg2+); error bars = sd.