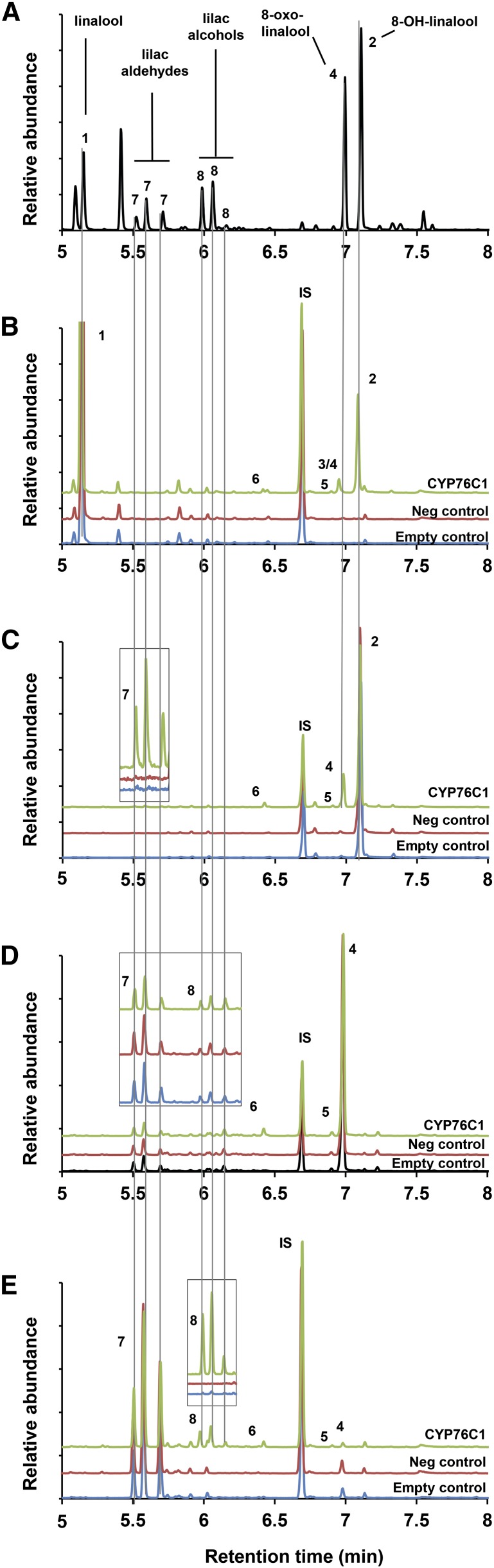
Figure 3.
Gas Chromatography Analysis of the Products Resulting from Yeast-Expressed CYP76C1 Activity on Linalool and Linalool Oxides.
GC-MS chromatograms of a mix of authentic standards (A) and of ethyl acetate extracts of the products from the conversion of a racemic mix of (R/S)-linalool (B), 8-OH-linalool (C), 8-oxo-linalool (D), and lilac aldehydes (E) by yeast-expressed CYP76C1. Microsomal membranes from the recombinant yeast expressing CYP76C1 (final [P450] ∼50 nM) or transformed with an empty vector (Empty control) were incubated for 15 min with 200 µM of substrate in presence or absence (neg. control) of NADPH. Chromatograms show the relative abundance of total ion current and the selected ions m/z 111 + 153 + 155 in the inserts. Compared with control, CYP76C1-dependent 8-oxo-linalool metabolism results in a decrease of 8-oxo-linalool and lilac aldehydes and a simultaneous increase in lilac alcohols (D). This suggested that the lilac aldehydes formed via conversion of 8-oxo-linalool might be converted rapidly into alcohols by CYP76C1 (E). Formation of lilac aldehydes from 8-oxo-linalool was confirmed performing a time-course experiment using low substrate and P450 concentrations (Supplemental Figure 7). (1) Racemic R/S-linalool, (2) 8-OH-linalool, (3) 9-OH-linalool, (4) 8-oxo-linalool, (5) 8-OH-6,7-dihydrolinalool, (6) 8-oxo-6,7-dihydrolinalool, (7) lilac aldehydes, and (8) lilac alcohols. IS, nonyl acetate used as internal standard for normalization. (2), (4), (7), and (8) were identified by comparison of RT and MS with those of authentic standards (Supplemental Figure 4). (3) was identified by comparison of its MS with MS of 8-OH-linalool (2) when separated from (4) on a HP-35ms column (Supplemental Figure 3). (5) and (6) were identified by NMR after reaction upscaling and purification by preparative GC (Supplemental Figure 6).