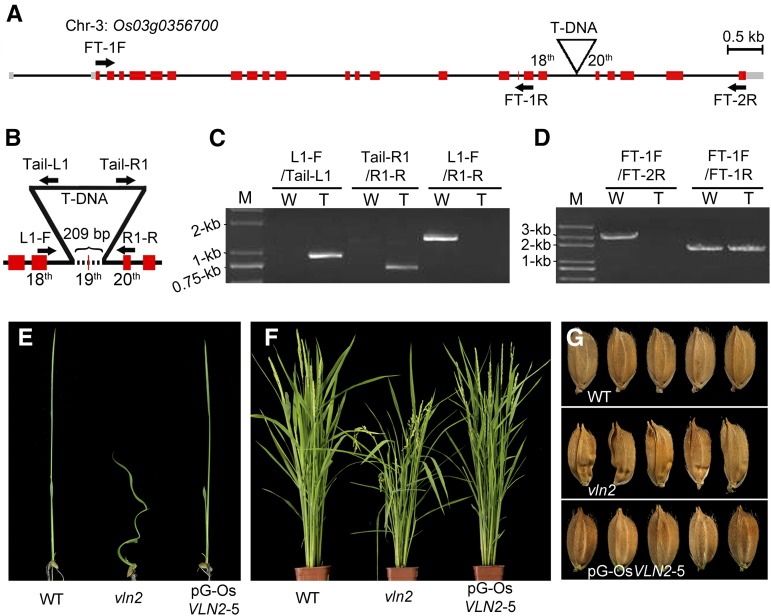
Figure 3.
Cloning and Genetic Complementation of vln2.
(A) Genomic structure of Os03g0356700 and the T-DNA insertion site on chromosome 3. Gray boxes, red boxes, and black lines indicate 5′ and 3′ untranslated region, exon, and intron, respectively. Arrows indicate the binding sites of primers used to detect the transcription of Os03g0356700 in the wild type and vln2 shown in (D).
(B) T-DNA insertion caused deletion of a 209-bp fragment (the dotted line), spanning the 19th exon and partial sequences of the 18th and 19th introns. Arrows indicate the binding sites of primers used to verify the T-DNA insertion site in (C).
(C) Confirmation of the T-DNA insertion site. Primers on both the genomic sequence and T-DNA were used and are shown in (B). W, wild type; T, the T-DNA insertion mutant vln2; M, molecular weight markers.
(D) The expression of Os03g0356700 is disrupted by T-DNA, leading to its incomplete transcription in vln2. The primers used are shown in (A). W, wild type; T, the T-DNA insertion mutant vln2; M, molecular weight markers.
(E) to (G) Complementation of the mutant phenotype by a 14.8-kb genomic vln2 sequence from the wild type, showing the restored seedling (E), flowering plant (F), and seeds (G). pG-OsVLN2, the construct containing a VLN2 genomic sequence including its promoter region.