Figure 1.
RNA-Seq Analysis of HD Isogenic Stem Cell Model
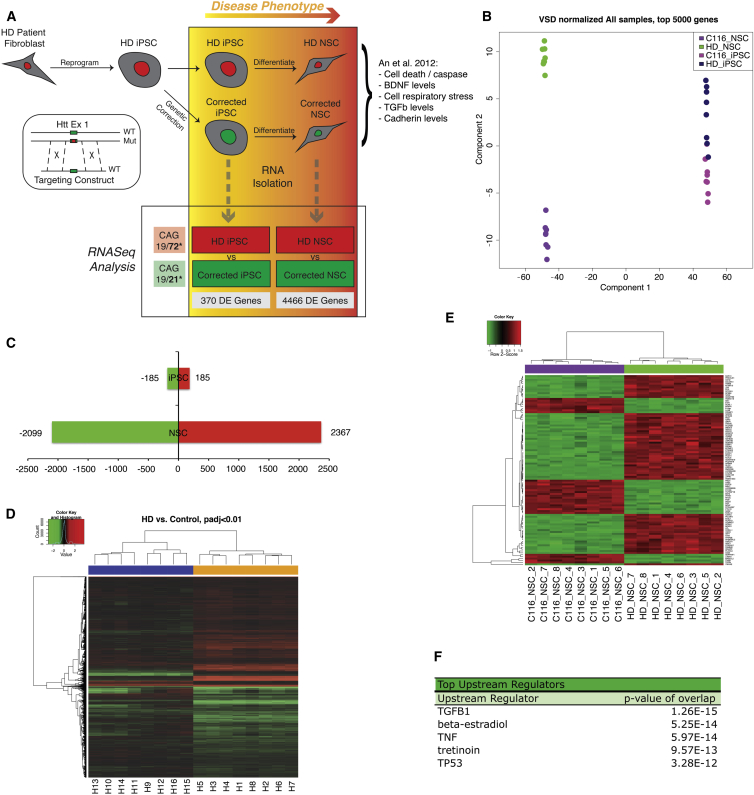
(A) Schematic illustration of experimental design and analysis. iPSCs reprogrammed from HD patient fibroblasts were genetically corrected for the HTT mutation using traditional homologous recombination via a targeting construct. Resulting isogenic iPSC pairs and differentiated isogenic NSC pairs were grown in tandem and plated as replicates for RNA isolation and RNA-seq analysis. There are eight biological replicates (BR) of each-corrected iPSCs, HD iPSCs, corrected NSCs, and HD NSCs: a total of 32 samples.
(B) Multi-dimensional scaling based on VSD-normalized expression levels of the top 5,000 most variable genes.
(C) Number of DE genes across comparisons at <0.01 false discover rate (FDR) threshold. Green bars represent the numbers of downregulated genes and red upregulated for each comparison.
(D) Heatmap representing DE genes in any of the comparisons of HD versus control. Samples (in columns noted H1–H16) and genes (in rows) are clustered by similarity. Shades of red represent upregulation, shades of green downregulation.
(E) Heatmap representing relative expression levels of the DE genes when comparing HD NSCs and corrected NSCs at an adjusted p value of <0.01. Gene names are listed on the right. The nomenclature is the NSC corrected line C116 and the sample number (C116_NSC_#) and HD-NSC and its corresponding sample number (HD_NSC_#).
(F) Table summary of top upstream regulators in HD versus corrected NSC samples via IPA analysis.