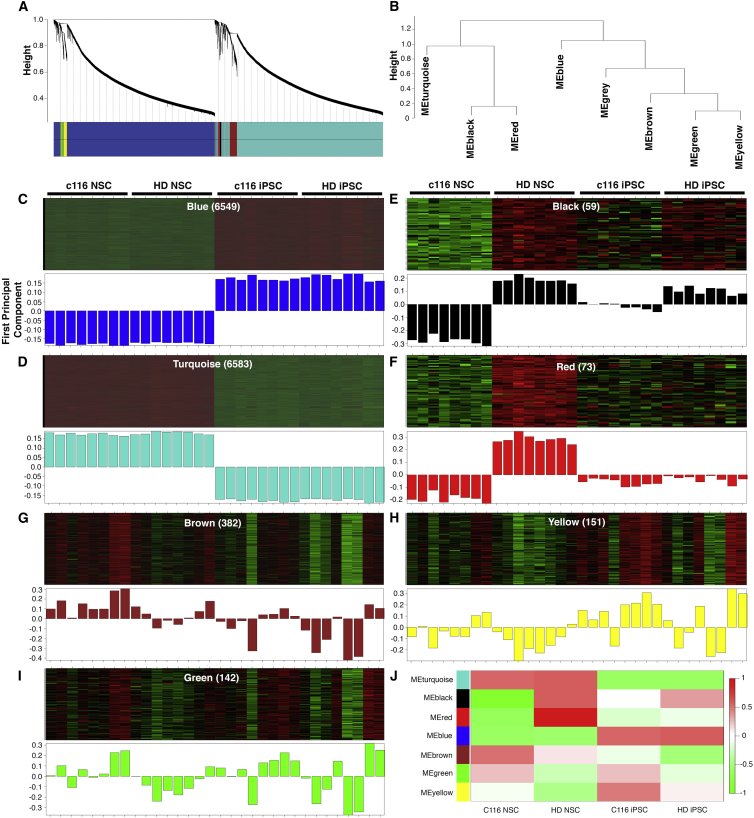
Figure 2.
WGCNA Analysis of HD Isogenic Stem Cell Model Reveals Drivers of HD Phenotypes in NSCs
(A) Dendogram of genes clustered based on topological overlap. Identified modules are defined by colors.
(B) Dendrogram representing the seven modules identified by WGCNA. The gray module represents genes that are not included in any of the modules.
(C–I) Heatmap (top) and first principal component (eigengene, bottom) of expression data for genes in WGCNA modules across 32 samples. Samples are labeled by group, and the number of genes in each module is in parentheses. Of seven distinct modules defined, turquoise and blue modules are characterized by coexpression traits consistent with expression differences between the iPSC and NSC state (C and D), whereas other individual modules define expression profiles consistent with changes in gene expression in NSCs due to CAG expansion (E and F) y axis is first principal component for (C)–(I).
(J) Summary of all modules by sample group.