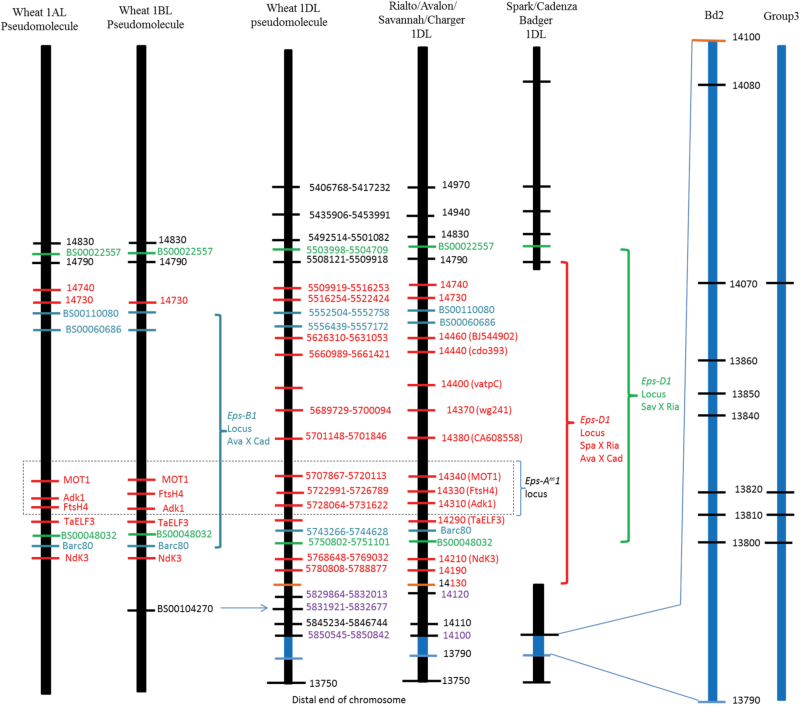
Fig. 2.
Schematic presentation of the Eps-B1 locus for Avalon X Cadenza (QTL confidence interval between SSR marker XBarc80 and KASPar markers XBS0011080 and XBS00060686, Eps-D1 (deletion of several genes on 1DL of Spark), Eps-D1 for Savannah X Rialto (no deletion) QTL confidence interval between KASPar markers XBS00048032 and XBS00022557. The dotted rectangle marks the Eps-Am1 locus and the two proposed candidates MOT1 and FTSH4 (Faricelli et al., 2010). The blue and green horizontal numbers and letters indicate SSR marker (XBarc80) and KASPar markers beginning with BS. The black vertical bars indicate the 1AL, 1BL, and 1DL wheat pseudomolecules from the wheat (IWGSC-based) pseudomolecule v3.3 (JIC) database and the region is collinear with Brachypodium chromosome 2. The red or black numbers on the right of Rialto 1DL next to the red or black horizontal bars, respectively, indicate the Brachypodium chromosome 2 gene numbers syntenic with wheat 1DL. The red numbers and letters on the right of Rialto/Avalon/Savannah/Charger 1DL indicate the genes deleted from Spark while the black numbers show those outside the 1DL deletion. The purple coloured numbers on the right side of wheat 1DL psuedomolecule and Rialto/Avalon/Savannah/Charger 1DL denote the genes between TaBradi2g14130 and TaBradi2g13790 that have not been checked if they are in the deletion but are likely to be outside the 1DL deletion. The gap on the vertical bar labelled Spark/Cadenza/Badger 1DL indicates the 1DL deletion. The blue vertical bars (Bd2 and group 3) are used to illustrate that all the eight brachypodium genes (13800, 13810, 13820, 13840, 13850, 13860, 14070, and 14080) between the predicted genes Bradi2g13790 (designated 13790 on Fig. 2) and 14130 do not match the wheat group 1 genes but four of these genes match the group 3 chromosome genes. The gene TaBradi2g14130 (14130) is labelled in black and red because part of this gene was amplified and sequenced from Spark while the rest of the gene seems to be deleted.