Figure 4.
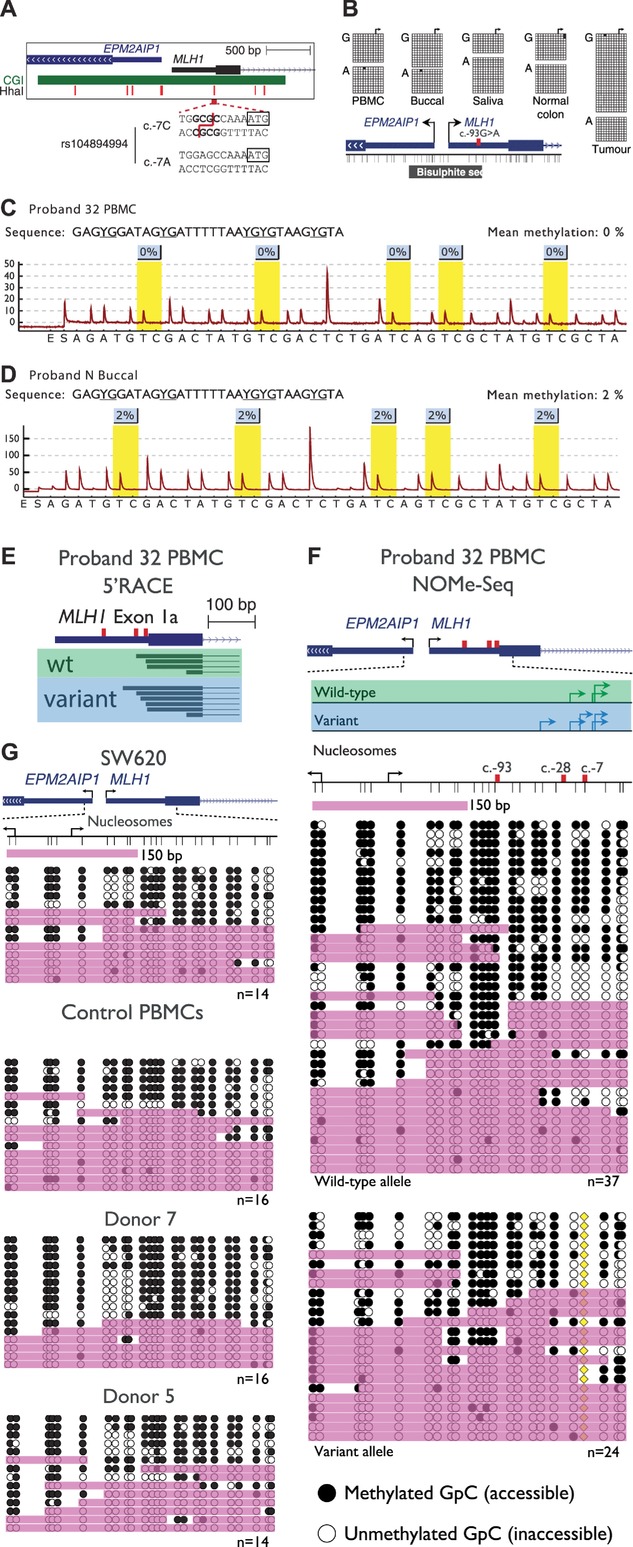
The variant allele is not epigenetically altered at the MLH1 promoter. A: A schematic of the MLH1 and EPM2AIP1 bidirectional promoter indicating the location of the CpG island (green bar), seven HhaI sites used to detect methylation by MS-MLPA (red vertical bars), and the sequence encompassing the c.-7C>T site. The presence of the c.-7C>T variant abolishes a HhaI restriction site. B: Single molecule bisulfite sequencing data of various tissues from Proband 32. The c.-93G>A site was used to distinguish between wild-type (c.-93G) and variant (c.-93A) MLH1 alleles. The black horizontal bar labeled “Bisulfite seq” indicates the region analyzed. Both alleles were unmethylated in all tissues examined including tumor tissue. C and D: Representative pyrograms indicating methylation levels at five CpG sites within the MLH1 promoter in Proband 32 PBMCs and Proband N buccal DNA, respectively. The nominal limit of quantification for this assay is 5%. E: The locations of unique transcription initiation sites in exon 1a of the wild-type (green box) and variant (blue box) MLH1 alleles. The c.-93, c.-28, and c.-7 sites are indicated by the red vertical bars. F: Nucleosome occupancy across individual promoter molecules separated according to allele of origin, as determined by the c.-7C>T variant (yellow diamond). Black arrows indicate the annotated MLH1 [NM_000249.3] or EPM2AIP1 [NM_014805.3] transcription initiation sites, whereas green and blue arrows indicate the locations of sites identified by 5′RACE in wild-type and variant alleles, respectively. Thin vertical black lines represent the positions of GpC dinucleotides. Black circles = GpC dinucleotides methylated/accessible to the GpC methyltransferase M.CviPI. White circles = GpC dinucleotides unmethylated/inaccessible to GpC methyltransferase. Pink shading indicates regions of accessibility ≥150 or >75 bp at the extreme ends of amplicons. G: Nucleosome occupancy across the same region in MLH1-expressing colorectal carcinoma cells and PBMCs from healthy donors. The number of molecules sequenced is indicated at the bottom right of each panel.
