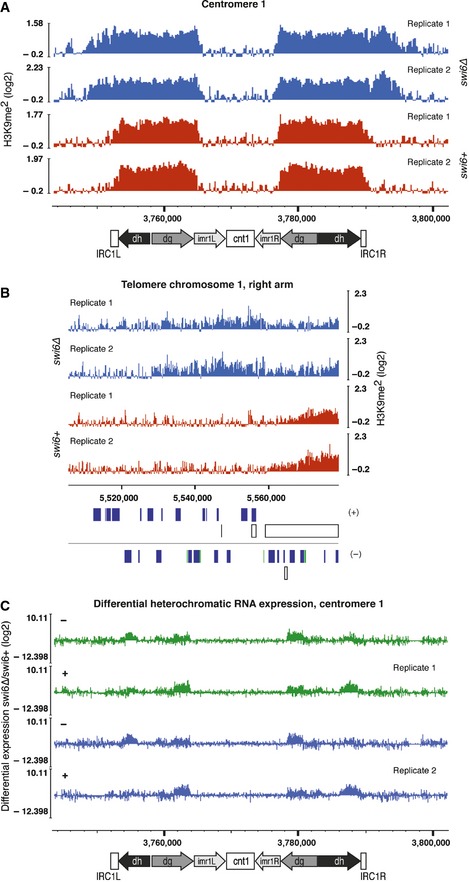
H3K9me2 ChIP‐seq enrichment profiles for
swi6+ and
swi6∆ cells on centromere 1. Centromeric repeat elements (dg/dh) are indicated. Inverted repeats (IRC1) constitute heterochromatin boundaries in wild‐type cells (Cam
et al,
2005). The
y‐axes represent log
2 ChIP‐seq enrichments in 200‐bp genomically tiled windows calculated over
clr4Δ cells. Two independent biological replicates for each genotype were performed (replicates 1 and 2). Positions of genomic elements on the plus and minus strands are indicated. Blue, protein‐coding genes; white, repeats; green, long terminal repeat (LTR).