Figure 2. Large macromolecular complexes of complement proteins assembled upon complement activation.
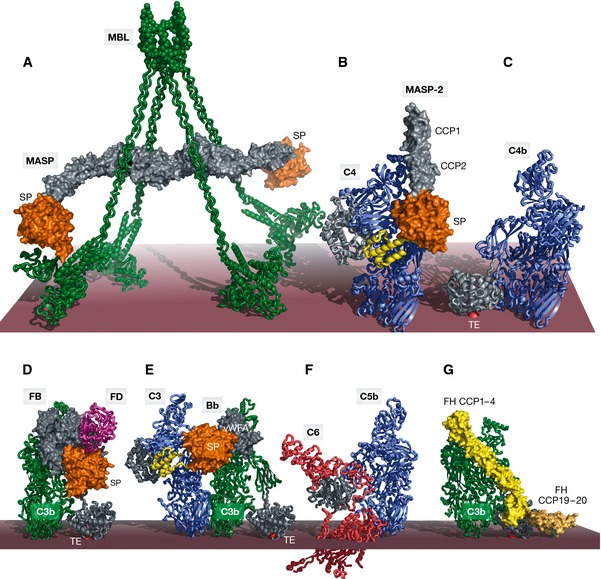
The order of panels (A–F) reflects the order of appearance starting from activation in the LP and ending with MAC assembly in the TP. (A) SAXS model of the MBL:MASP‐1 complex with MBL (green) associated with a MASP‐1 homodimer with its serine protease domains (orange) protruding away from the MBL collagen stems in agreement with an intercomplex activation mechanism. (B) Crystal structure of the C4:MASP‐2 complex (RCSB ID 4FXG) with the substrate (C4, blue with the anaphylatoxin domain in yellow) making contacts at two distinct sites; the CCP domains (gray) and the SP domain (orange). (C) Crystal structure of C4b (RSCB ID 4XAM) with the TE domain colored in gray and the reactive thioester covalently bound to the membrane shown as a red sphere. (D) Crystal structure of the ternary C3bB:D complex (RSCB ID 2XWB). FB binds C3b (green, with the TE domain in gray) via its vWA and 3 CCP domains (gray). The SP domain (orange) is in the closed state. FD (magenta) is recruited to FB. (E) Structural model of the AP C3 convertase in complex with a C3 substrate (blue) generated by superimposing C3bBb stabilized with SCIN (RCSB ID 2WIN) and the C5:CVF complex (RCSB ID 3PVM). The anaphylatoxin moiety (yellow) is released upon cleavage. (F) Crystal structure of the C5b6 complex (RCSB ID 4A5W) revealing conformational rearrangements occurring upon C5 cleavage to C5b (blue), reminiscent of those observed in the C3/C4 to C3b/C4b conversion. (G) Structural model of FH binding to C3b (green) generated by superimposing C3b bound to FH CCP1–4 (light yellow, RCSB ID 2WII) and TE domain bound to FH CCP19–20 (dark yellow, RCSB ID 4ONT), CCP5–18 are not illustrated. FH also interacts with host glycans. The binding of FH prepares C3b for FI binding and cleavage. In all panels, the red surface approximates the activator such as the surface of an LPS layer on a pathogenic bacterium. Importantly, this is separated from the cell membrane; thus, panel (F) does not imply that C6 extends into the membrane.
