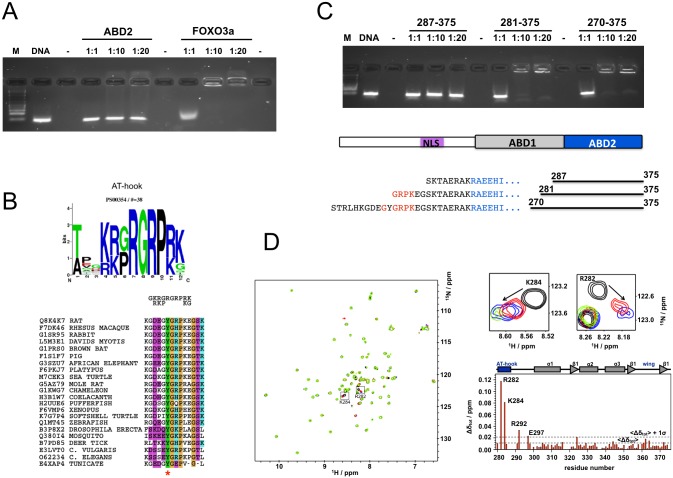
Fig 2. DNA binding of Ms1.
A: EMSA experiment of ABD2 and FOXO3a as a positive control with a random 18mer library (2 μM). B: consensus motif of the AT-hook shown as web logo [40] (based on PROSITE entry PDOC00306) and in short form on top of a sequence alignment of the AT-hook motif in selected Ms1 sequences. Colours are: purple—hydrophilic; green—hydrophobic; orange—proline and glycine; blue—positive charge. Phosphorylation sites identified experimentally are indicated by a star [15,41]. C: EMSA of N-terminally extended ABD2 constructs with the same 18mer random library used in A. The overall Ms1 topology is shown together with the detail of the N-terminus of the various constructs. Residues at the N-terminus of ABD2 are shown in blue and those of the AT-hook in red. D: NMR analysis of the interaction of Ms1 270–375 with AT-rich DNA ds oligonucleotides. Spectra of Ms1 270–375 without (black) and with (1:0.5 red; 1:1 blue; 1:2 green) of the oligonucleotide are shown superimposed together with a detailed view for two selected residues from the N-terminus. The chemical shift perturbations are summarised in a plot against the amino acid sequence that is annotated with the functional and structural properties of the 270–375 construct.