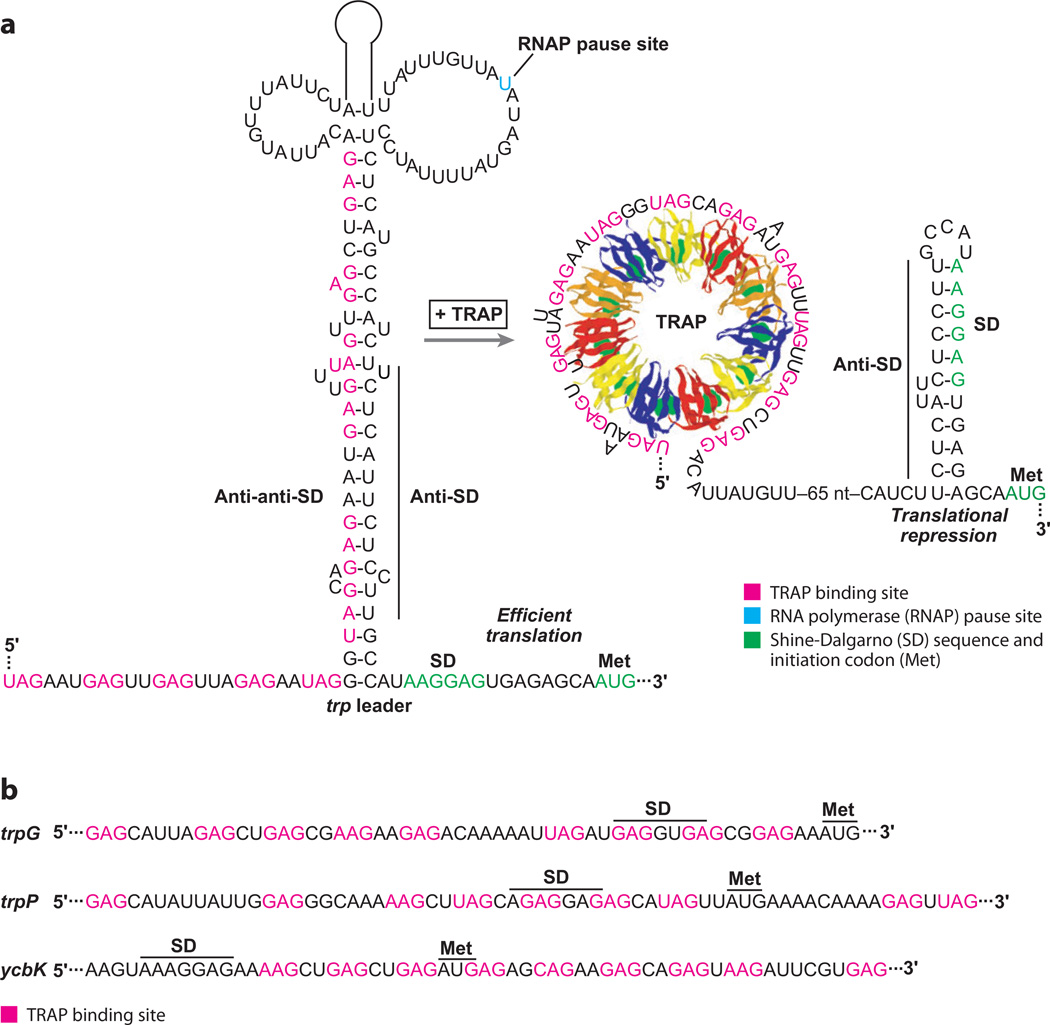
Figure 4.
Translational repression of Bacillus subtilis tryptophan metabolism genes. (a) Model of the trpE translational repression mechanism. RNA polymerase pausing during transcription provides additional time for binding of tryptophan-activated trp RNA binding attenuation protein (TRAP). In the absence of bound TRAP (limiting tryptophan), the RNA adopts a structure such that the trpE Shine-Dalgarno (SD) sequence is single stranded and available for ribosome binding. In the presence of excess tryptophan, TRAP binding promotes formation of the trpE SD-sequestering hairpin. (b) TRAP binding sites in the trpG, trpP, and ycbK transcripts. In each case, bound TRAP competes with ribosome binding and represses translation. The SD sequence and initiation codon (Met) for each mRNA is marked.