Abstract
Data sharing and mediation across disparate neuroimaging repositories requires extensive effort to ensure that the different domains of data types are referred to by commonly agreed upon terms. Within the SchizConnect project, which enables querying across decentralized databases of neuroimaging, clinical, and cognitive data from various studies of schizophrenia, we developed a model for each data domain, identified common usable terms that could be agreed upon across the repositories, and linked them to standard ontological terms where possible. We had the goal of facilitating both the current user experience in querying and future automated computations and reasoning regarding the data. We found that existing terminologies are incomplete for these purposes, even with the history of neuroimaging data sharing in the field; and we provide a model for efforts focused on querying multiple clinical neuroimaging repositories.
Keywords: Neuroimaging, data sharing, clinical scales, assessments, mediation
1 Introduction
Using magnetic resonance imaging (MRI) in cognitive neuroscience and neuropsychiatry has resulted in decades of study-specific datasets being stored at various research institutions or in warehouses of archived data [1]. These data may or may not have been used in a publication, or even analyzed; however, they can in many cases be combined in new analyses, or re-examined with new methods for new findings. They form an investment in brain images and information that needs to be capitalized upon.
As a result of the neuroimaging community’s growing awareness that MRI datasets can and should be shared for accelerating scientific discovery, a large number of repositories have been developed and made available. Recent developments on data harmonization have led to the creation of national databases such as the National Database for Autism Research (NDAR) [2]. Within the imaging community studying schizophrenia, it was recognized that large scale datasharing would encourage reproducibility, generalizability, and special analyses of rare subjects [1, 3]. The data repositories developed by the Functional Imaging Biomedical Informatics Research Network (FBIRN; [3–5]), by the Mind Research Network (MRN) [6, 7], and the XNAT Central project [8–10], all included schizophrenia research imaging datasets, with the associated clinical and subject-specific information. These repositories were all developed with an eye toward solving the problem of data sharing: the FBIRN system, the Human Imaging Database, HID, is a federated system that allows the same database to be installed and queried across various collaborating institutions. It has a userbase of about 25, with several thousand downloads (D.B. Keator, 2015, personal communication). The database was carefully designed to be extensible and generalizable to archive clinical, imaging, and any other data type from any sort of study. The MRN system, the Collaborative Imaging and Neuroinformatics System or COINS, also includes a complex but extensible relational database to both archive data and manage ongoing projects, with additional tools for importing images and linking to imaging pipelines, anonymizing data on the fly for sharing, managing data sharing requests, etc. Including both data providers and data users, it has a userbase of over 1300 unique users in 38 states and 34 countries around the world (http://coins.mrn.org/index.php?page=userMap). The XNAT Central system is a lightweight data management system primarily for archiving and sharing imaging data from a variety of studies; it has a userbase in over 100 different institutions, each with approximately 50 users (D. Marcus, 2015, personal communication).
The SchizConnect project (www.schizconnect.org) [11] was developed to connect these and related imaging repositories so that a single query, e.g. for the data from all male subjects with schizophrenia and a DTI scan who have some measure of executive function, could return information from all the available schizophrenia imaging repositories. In these three example repositories noted above are data from several hundred patients and an equal number of control subjects from several different studies (for a total of 1091 subjects as of the time of writing). The data types per subject included the imaging data from structural and functional imaging, the subject specific demographics such as age, gender, diagnosis, and other measures, the subject’s scores on clinical scales regarding various symptom profiles, and the subject’s scores on cognitive test batteries. Each study in the various repositories had its own design, with its own choice of variables and scanning data for each subject. In some repositories, the imaging and clinical data are kept in separate databases with linking IDs; in others there are very stringent access rules to data, with complex layers of approval for any query that may vary with the study being queried. The details of SchizConnect’s mediation system to solve this problem are presented in a companion paper. In this paper we describe the work we have done in harmonizing the terms used across the different sources and studies.
There are at least two usages of “harmonization” that come up in this project. The first is harmonizing data from different studies so that a data point from one study means the same thing as that data point in a different study from a different research team. We know, for example, that different MRI machines do not create identical pictures of the same brain [12–14]; different machines will provide unique regional contrast values across tissue types, and different imaging protocols will introduce specific distortions in the image. While cognitive neuropsychology tests are often harmonized, so that for example, an IQ of 100 is roughly comparable regardless of the specific standard IQ test, and clinical scales are standardized so that for a given scale neuropsychiatrists know what a score of 0,1,2, etc. should mean for the severity of the subject’s symptoms, it turns out that without careful calibration of the observer or clinician, the same subject with the same clinical interaction may receive a different value from different raters. “Harmonizing” the data in this case means taking into account that both the people and the machinery used to collect the data introduce a bias or effect which is different from study to study, and harmonization methods remove that to make the data more directly comparable across sources. The best methods for taking this variation into account are not always known, and are outside the scope of SchizConnect.
The second meaning of “harmonization” is much simpler, on the one hand, but much more basic to the aims of datasharing, on the other. In building data repositories, many decisions are made that are specific to that particular repository or study, about what they will call different datatypes. The mediation efforts include implementing queries to each data source, so that the general user’s query can be translated into a query that will retrieve the right data from each database regardless of differences in the database’s structure. While the bulk of that work is in dealing with the structural differences in the database models, there are terminology differences which also need to be solved. In one study’s data a structural MRI scan may be listed informatively as “T1-weighted scan”, or something as complex as “5MPRAGE-AVG” or just “scan1”, which assumes someone knows that to get the T1-weighted structural images they should look in “scan1”. Harmonizing the data in this case means mapping the terms to standard terms that capture the semantics of what the data actually are, to help the user and eventually automated systems find the right data.
Lists of standard terms with definitions and uniform resource identifiers (URI) are often described as ontologies. Technically, a fully-developed ontology also includes logical definitions and relationships among the terms, rather than just a terminology list [15]. However, many ontologies or simpler lexicons have been published and shared for general use either with or without the more rigorous logical definitions, with the goal of providing standard terms that can be referred to by semantic web technologies. Ideally, within SchizConnect the terms being used for harmonization would also be standardized, with clear definitions and permanent URIs, so that there is less ambiguity both from the human user and from eventual automated systems when performing queries across resources.
Thus our goals in this part of the project were to develop three terminologies for the multiple data domains available across the resources: 1) imaging types, 2) cognitive measures, and 3) clinical variables, focused on the schizophrenia datasets. We first identified what the needed terms were, identified a basic data model for each domain, and examined the available ontologies and terminology resources for possible standardized terms. In many cases the existing terminologies were not adequate, which entails development and dissemination of new terms. This project builds on many previous efforts, and provides a research-oriented integration of several different facets in service of a single endeavor, as an example that can be leveraged in turn for other similar projects. We describe the needed steps and specific issues we faced; the specific terms and definitions are available for download from SchizConnect.org.
2 Methods
2.1 Identifying the needed terms from sources
We extracted the database-specific terms from the different source repositories, and identified the different terms used for the same datatypes. Each data repository team provided a list of the variable names that could be queried, broken up into whether they referred to imaging data, or other variables. The terms were then compared, to identify which terms were actually referring to the same thing, or different things. This required extensive human interaction across teams, to identify when variable names were being used consistently both within and across repositories. The expertise needed for this effort included both the study-specific information from data collectors, database designers, and the domain expertise from neuroimagers and neuropsychologists.
A key issue in determining terms and definitions is to consider the granularity of the queries: Identifying that a subject has a particular standardized image type or clinical variable is one level, and that is the level that SchizConnect is focused on facilitating in this initial development. On the other hand, querying based on what the measure is about or what it is supposed to measure is a very different level of granularity. Many data points are actually composite, in that they are sums of measures on different questions about a subject’s level of social function, for example; querying whether there is a measure of anxiety included on any test available in the repository requires a fine-grained semantic modeling which is not yet available through SchizConnect. Similarly, the functional MRI studies include cognitive behavioral tasks collected during the scan, measuring cognitive processes such as working memory or auditory processing; querying whether the fMRI data includes experimental conditions that entail specifically visual working memory, for example, requires an infrastructure that we want to be able eventually to include in our modeling.
2.2 Mapping the source terms to a domain model
Once the variables were identified and roughly defined, we then identified the domain model, or the hierarchy of terms for each of our three domains (imaging, clinical, or cognitive neuropsychological measures). In order to determine the hierarchy we compared our models with existing ontologies, and with the understanding of the relationships among the terms and models that the userbase for SchizConnect had.
For each term that we included in the domain model, we then identified the definitions of each term, mapping to other source ontologies when possible. We chose to use several established sources, namely UMLS (http://www.nlm.nih.gov/research/umls/), SNOMED (http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html), NIFSTD/Neurolex [4, 16, 17], and Cognitive Atlas/Cognitive Paradigm Ontology (CogPO; http://www.cognitiveatlas.org/ and http://cogpo.org/) [18, 19]. We also searched Bioportal.bioontologies.org [20] for potential matches, as that simultaneously searches several hundred biomedically relevant ontologies. However, we prioritized the ontologies listed previously as sources of terms, since not all ontologies that have been published are either complete or being actively maintained.
2.3 Build the terms into the Mediator and Query Portal
The primary use of these terms in the current instantiation is for human users, to facilitate their understanding of how to query for what they might want. Thus these terms form the basic vocabulary for querying SchizConnect. As the hierarchies are developed, the querying interface develops to incorporate them, and the mediator system uses them and mappings to the terms in the sources to build the executable queries sent to the data sources. The details of how this is done are presented in a companion paper by Ambite et al. on the SchizConnect mediator.
3 Results
The spreadsheets of the different terms, their hierarchical structures and definitions are available for viewing and download at schizconnect.org. The terms are in the process of being submitted to Neurolex (www.neurolex.org) when Neurolex URIs do not already exist. The spreadsheets as current working drafts are available at http://schizconnect.org/documentation#data_models.
3.1 Imaging hierarchy
Collecting all the specific variable names for the imaging sessions across the different repositories, we identified 632 idiosyncratic labels (e.g., “ep2d_words” for a particular task-based fMRI scan, “MR-010” for a structural scan). In order to find all the T1-weighted images that could be used to extract brain volumes from the COINS repository, for example, one needed to know that across all the available studies there were 29 different strings that labeled that kind of image. Our final, harmonized list currently consists of 22 unique terms, described generally below.
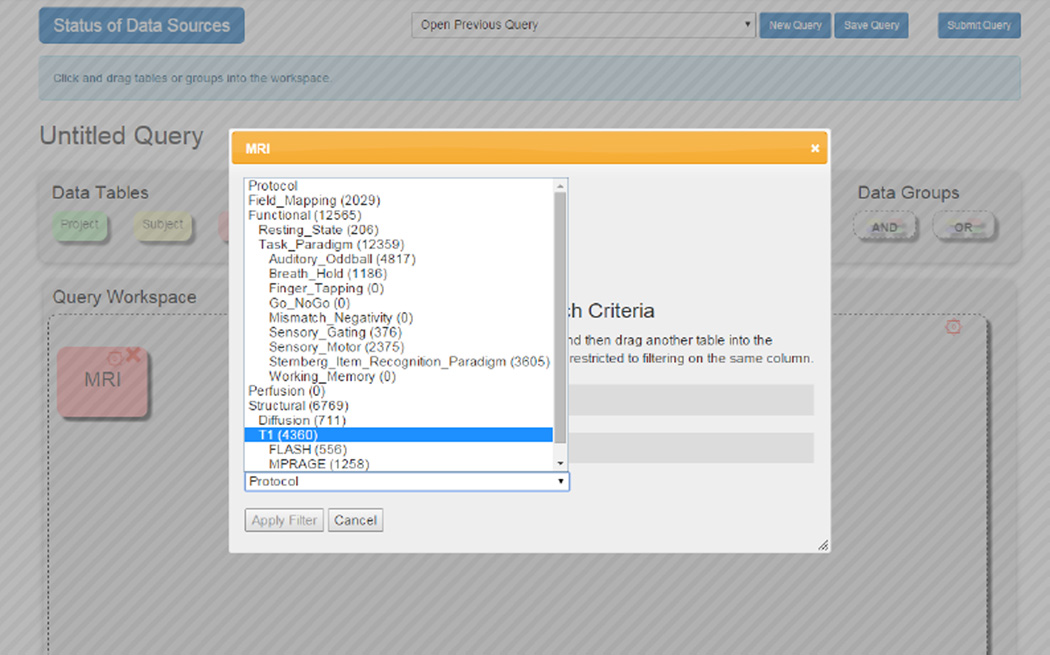
We modeled the original imaging labels as referring to several basic types of imaging data: Structural, Functional, Fieldmapping or Perfusion. Every imaging series that is collected can be in only one of these categories. Structural scans measure the anatomy of the brain, and under Structural scans we included T1, T2, and Diffusion. (See Figure 1.) These are shorthand for, respectively “3D T1-weighted scan”, or nlx_inv_20090243 from NeuroLex; “T2 weighted MRI 3D image”, or nlx_156812; and “Diffusion weighted MRI 3D image”, or nlx_156811. Functional scans are also referred to as functional MRI or fMRI, and measure the Blood Oxygenation Level Dependent (BOLD) signal changes. This label is defined as “Functional MRI Assay” or nlx_inv_090914 from NeuroLex. Perfusion scans include Arterial Spin Labeling (ASL) scans, which measures the flow of blood through the brain, generally speaking. Fieldmapping are scans collected specifically to measure distortion in the magnetic field. Neither of these terms had matches in NeuroLex. The functional MRI scans were separated by “resting state” or “task-based”, and if task-based, what the task was. The task could often be linked back to a pre-defined term in CogPO or Cognitive Atlas.
Fig. 1.
Example of the Imaging Hierarchy being used in the Query portal for SchizConnect.
This hierarchical structure specifically reflects the research community needs; it is very different, for example, from the hierarchical structure for RadLex [21, 22]. We decided on function or intent of the scanning protocol as the basis for categorization, rather than the imaging parameters per se. Radiologists and MRI physicists would organize the scanning types very differently, based on exactly what the scanning sequence parameters and details were. In our case, not all T2-weighted scans are structural; a T2-weighted scan that was used to measure some marker of brain function would be classified under “Functional.” However, within the structural images, distinguishing a T1-weighted from a T2-weighted image is very important for analysis purposes and thus is modeled explicitly.
The choice of labels of “Structural” or “Functional” is shorthand for the benefit of the cognitive neuroscience or neuropsychiatric research community, who look for images that they can use to identify brain measures reflecting anatomy or physiology. This is very similar to the structure identified separately in the Quantitative Imaging Biomarker Ontology (http://purl.bioontology.org/ontology/QIBO) [23], which also explicitly breaks imaging measurements into “Anatomical” and “Functional” classes.
3.2 Neuropsychological assessments hierarchy
There were several standard cognitive batteries included with the various datasets, which overlapped in what they measured (attention, memory, verbal fluency) etc., but not in the particular test used. In consultation with neuropsychologists, we identified 11 subdomains, each of which had several specific tests or test modules which measured it. Examples are shown in Figure 2 below. Specifically, under measures of “Verbal Episodic Memory”, the available datasets included scores from several standardized tests of immediate or delayed recall and recognition. Overall, we began with 67 neuropsychological tasks terms across the different datasets and reduced it to 49 common tasks at the most granular level. Many of the general domains as well as specific tests had terms with URIs from Cognitive Atlas, rather than SNOMED or other sources.
Fig. 2.
Part of the Neuropsychological assessment hierarchy for querying in SchizConnect. The number of subjects with data from each assessment are included in parentheses to help users identify the most common data types.
3.3 Clinical hierarchy
Within the Clinical section we included the Subject Types and measures specific to aspects of disease. We started with approximately 70 idiosyncratic terms and reduced that to 55.
Given the datasets we were harmonizing worked primarily with studies of people with schizophrenia or healthy control subjects, the list of subject types was expected originally to include two terms: schizophrenia or control. That however did not fit the reality of the datasets. Some inclusion and exclusion criteria were different across datasets: Some included only subjects who strictly fit the definition of schizophrenia with no previous different diagnoses; other studies were more broad and allowed subjects with schizoaffective disorder. The “control” samples were even more heterogeneous, in that each had their own exclusion criteria and others were more lax, requiring only no history of clinical psychosis. The one aspect that could be agreed upon for the “control” subjects was that they had no known or listed diagnoses at the time of inclusion at the study. There is no guarantee across all studies that they were healthy from the point of view of their cardiovasculature, occasional illicit drug use, exercise or sleep habits, for example, since screening and exclusion criteria were study-specific.
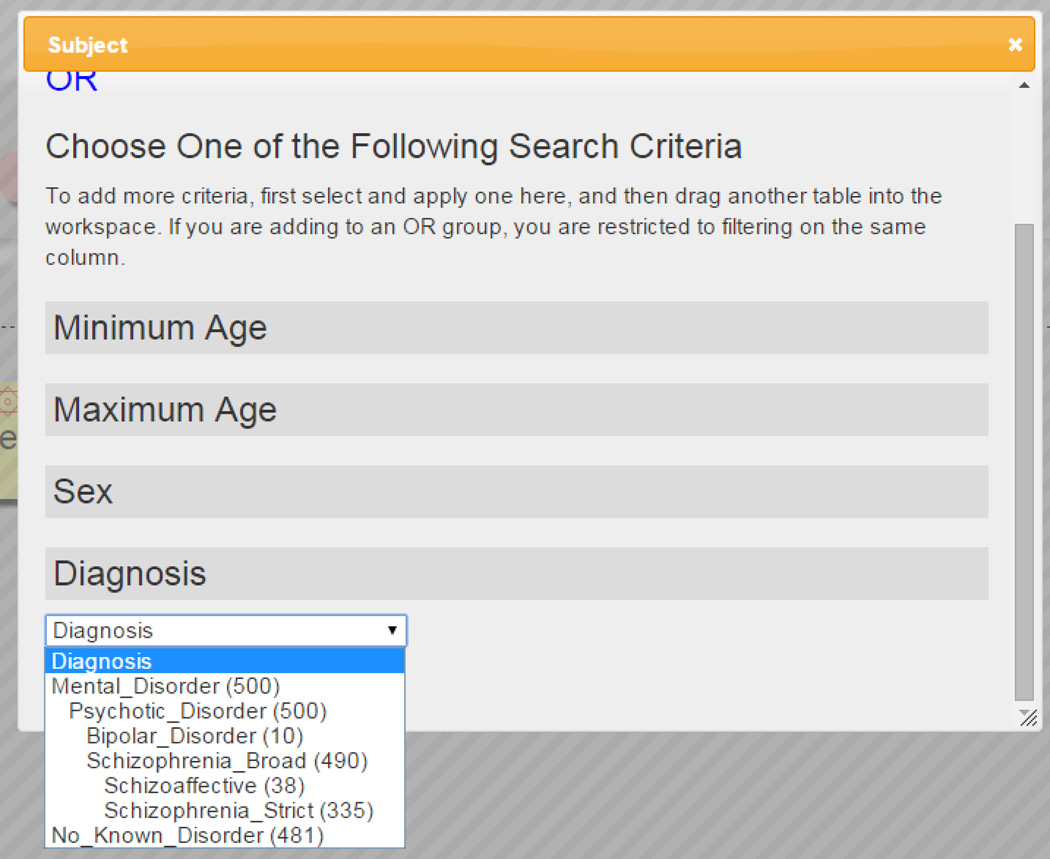
Thus the hierarchy under “Diagnosis” included: either “Mental Disorder” or “No Known Disorder”; under Mental Disorder was included “Psychotic Disorder” (allowing for multiple diagnoses later perhaps including non-psychotic disorders); as subclasses of Psychotic Disorder, both “Bipolar Disorder”, and “Schizophrenia (Broadly defined)”; then as subclasses of Schizophrenia (Broadly defined) were strict “Schizophrenia”, and “Schizoaffective”. See Figure below. This terminology is in principle expandable to include specific terminologies such as the ICD10 codes, or DSM-V codes, but that is not the researchers’ data. Specific diagnostic codes were not available, only whether a person fell into one of two groups: cases or controls.
Symptom severity measures and other clinical measures draw largely from standardized, published scales that fall into specific classes based on what they measure. We identified 14 subdomains or aspects of disease measured in these studies, such as “Extrapyramidal symptoms”, “Structured Interviews for Diagnosis”, or “Mood,” most of which had several scales used across the different studies. These classes do not have matches in any of the ontology sources we have examined to date; the standardized assessments largely can be pulled from SNOMED.
However, there were also idiosyncratic questionnaires to be included, such as specific post-imaging questionnaires assessing whether scanning exacerbated specific symptoms. That particular questionnaire may never be used again by another imaging study, but making it available through SchizConnect lets other researchers know it is there, leading them possibly to collect the same data, and more assessments may fall into that class in the future.
3.4 Evaluation
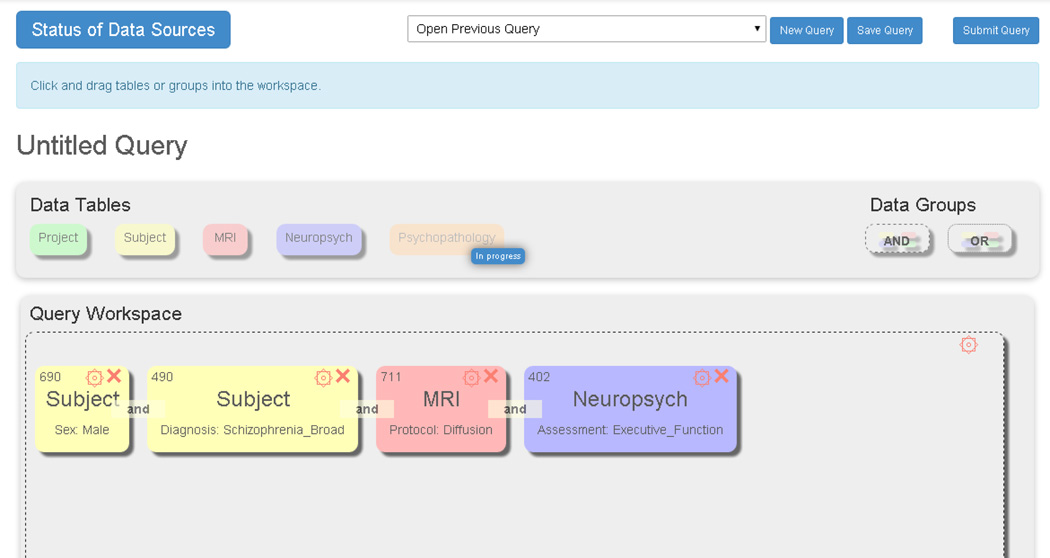
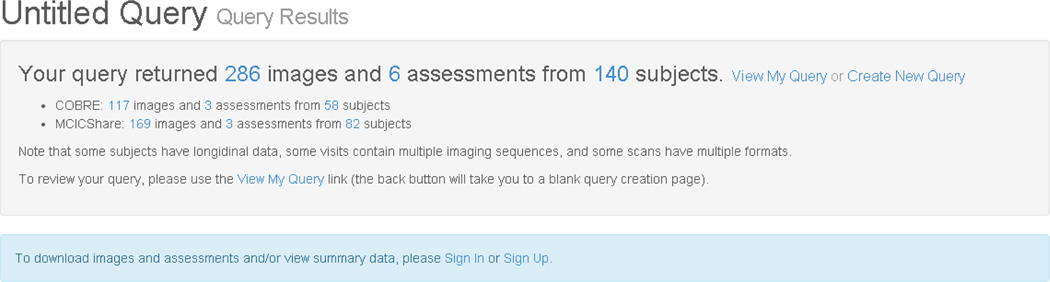
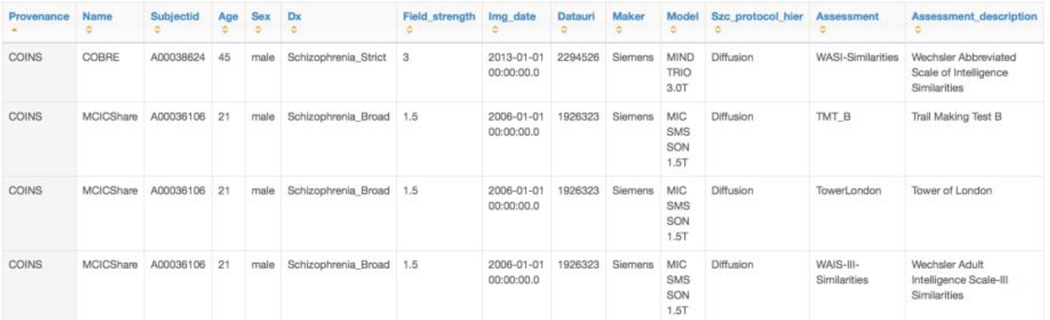
The SchizConnect portal has incorporated these terminologies in the querying capacity as shown above. Examples are shown in Figures 4, 5, and 6. An example final query is below, showing a request for male subjects with broadly-defined schizophrenia and a DTI scan who have some measure of executive function. The numbers of subjects meeting each filter is given in the upper left of each square box. The interface is a drag and drop one, based on the current Data Exchange interface from COINS. [24] The result in Figure 5 is the number of imaging datasets from how many unique subjects available across the various repositories; in this case, 286 images from 140 subjects. The users can then proceed to request the data or go back and modify their query. After signing the appropriate data sharing agreements, the user can also obtain the individual-level data, an excerpt of which appears in Figure 6.
Fig. 4.
Example query in Schizconnect, using the standardized terms.
Fig. 5.
The results of the query from Figure 4. The user can then proceed to request the data from the different repositories.
Fig. 6.
An excerpt individual-level results of the query from Figure 5. To obtain individual level results the user needs to sign the appropriate data sharing agreements.
Given the hierarchy we have included in the terms, investigators can query for subjects who have data at any level—requesting subjects who have any data on their executive function, for example, will return currently 402 subjects who have data from any of a number of cognitive tests. Or the query can drill down for only the subjects with data from the TrailMaking Test-B (TMT-B), to maximize comparability in the resulting dataset.
4 Discussion
Even with decades of work in the research community developing ontologies and terminologies to facilitate common communication across data repositories, we have identified several issues with the existing resources. It is simply not the case that we can identify the needed term for any given variable in any given clinical neuroimaging study from the work already done in UMLS or SNOMED or other sources. In this work, out of almost 200 terms needed, fewer than 50 have already been defined and given URIs, and the rest need new terms. This work of harmonizing terms across repositories continues to be largely manual, although the goal eventually is to automatically map new terms to known terms as new repositories are integrated.
Given the close collaborations between NIF and other ontology developers, both Cognitive Atlas and CogPO terms have Neurolex IDs. We chose to use the Neurolex IDs and include the original terms as synonyms. This was not an issue for UMLS and SNOMed as the overlap between them and other sources was much less. This leads to the different terms used in SchizConnect having different source ontologies, which may lead to issues in the future for automated reasoners, given the lack of logical rigor in many of the sources. This will be an ongoing part of the work, to have the SchizConnect data models all in a computable form and the terminologies released as well formed RDF/OWL files. Currently, the imaging model is under discussion with the International Neuroinformatics Coordinating Facility Data Sharing Task Force (INCF), as a basis for their OWL representations capturing terms and definition standards for imaging scan types. The cognitive and clinical models can be coded as OWL files in the future.
We did not use Common Data Elements (http://www.nlm.nih.gov/cde/) as a source of terms. Common Data Elements address a problem common to data sharing, that different studies use different data collection questionnaires, scales, and assessments. The CDE effort for many biomedical research domains is attempting to identify a minimum common core of measures to collect, and tools with which to collect them. Thus CDEs are often just pdfs of questions, not compatible with semantic web needs. Rather than define what an existing dataset’s assessments are, and represent the semantics in some way, they are proscriptive for future datasets. They reduce semantic uncertainty through providing a common set of measures, but not necessarily providing the semantic information regarding those measures. With the exception of the NINDS CDEs (http://www.commondataelements.ninds.nih.gov/CDE.aspx), there is a common lack of definitions and an overreliance on common usage, in the terms; URIs for individual terms are not always available; and they are often not available in an OWL/RDF format or other format which would allow extensions into computable representations of the terms, with automated reasoning available eventually. The NIH Toolbox, a set of cognitive assessments being recommended for use in clinical studies, was not used for any of the studies being modeled in the data repositories; if datasets which used the NIH Toolbox are accessed in SchizConnect in the future, we will assess the state of the relevant CDEs at that time. The CDEs are in ongoing development and will be integrated into the terminology usage whenever possible.
Common repositories of terminologies for clinical neuroimaging research are needed; UMLS is big, but not flexible enough for the day to day needs of modeling novel neuroimaging experiments where new variants of old concepts arise regularly. Bioportal [20] is useful as a repository of lexicons, for comparing across terminology sets to identify whether a term is already defined somewhere, and provides many tools for ontology-based data access; but in itself it doesn’t solve the problem of semantically representing what a given dataset of values mean, and what conclusions they can be used to support. The Ontology of Biomedical Investigation (OBI) is incredibly thorough and logically rigorous for the domains that have worked on it (vaccines, for example), but it requires expert effort to extend into new areas [25, 26]. It is in many ways the standard to aspire to, for supporting logical reasoning. NeuroLex [17] as a repository of terms is flexible, extensible by the community, and well-structured, which is at least the first step in aggregating a common set of standardized terms.
Finding the neuroimaging and associated clinical data is one aspect of mining and re-using neuroimaging data; using it is another. SchizConnect has focused on identifying datasets which fit certain high-level characteristics (gender, age, diagnostic group, scan type etc.). Other collaborative groups include the INCF Neuroimaging Data Model (NI-DM), which focuses on models of individual subject neuroimaging data collection, processing methods, and individual or group statistical analysis [27, 28]. The terms for these more detailed concepts also need to be shared in ways that make their definitions clear, at the very least, for re-use in other projects like SchizConnect. Currently SchizConnect cannot answer more nuanced queries such as “Find cognitive and imaging datasets that show gray matter loss in the anterior cingulate in adult patients with childhood-onset schizophrenia”, for example; one might be interested in the patterns of cognitive problems such patients have, and want to mine the available data to find out. With further development and interaction with the NI-DM development to represent gray matter loss analyses, and the Foundational Model of Anatomy (FMA) [29–31] to identify brain regions such as anterior cingulate cortex, such a query might be possible. This and other similar approaches being used in clinical research [32] would form the foundation for a truly innovative approach to large-scale, integrative biomedical science.
Fig. 3.
Diagnostic categories currently used in SchizConnect.
Acknowledgements
SchizConnect is supported by a grant from the National Institutes of Health (NIH/NIMH), 5U01MH097435 to L. Wang, JL. Ambite, S.G. Potkin and J.A.Turner. The work on COINS is also supported by 5P20GM103472 (NIGMS) to V.D. Calhoun. The authors would like to thank Derin Cobia, PhD, for help on constructing the SchizConnect neuropsychological assessment hierarchy.
Contributor Information
Jessica A. Turner, Email: jturner63@gsu.edu.
Matthew D. Turner, Email: mdturner46@gsu.edu.
David B. Keator, Email: dbkeator@uci.edu.
Kathryn Alpert, Email: k-alpert@northwestern.edu.
Margaret King, Email: mking@mrn.org.
Drew Landis, Email: dlandis@mrn.org.
Vince D. Calhoun, Email: vdcalhoun@mrn.org.
Steven G. Potkin, Email: sgpotkin@uci.edu.
Marcelo Tallis, Email: tallis@isi.edu.
Jose Luis Ambite, Email: ambite@isi.edu.
Lei Wang, Email: leiwang1@northwestern.edu.
References
- 1.Turner JA. The rise of large-scale imaging studies in psychiatry. GigaScience. 2014;3:29. doi: 10.1186/2047-217X-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall D, Huerta MF, McAuliffe MJ, Farber GK. Sharing heterogeneous data: the national database for autism research. Neuroinformatics. 2012;10:331–339. doi: 10.1007/s12021-012-9151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keator DB, Helmer K, Steffener J, Turner JA, Van Erp TG, Gadde S, Ashish N, Burns GA, Nichols BN. Towards structured sharing of raw and derived neuroimaging data across existing resources. NeuroImage. 2013;82:647–661. doi: 10.1016/j.neuroimage.2013.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bug W, Astahkov V, Boline J, Fennema-Notestine C, Grethe JS, Gupta A, Kennedy DN, Rubin DL, Sanders B, Turner JA, Martone ME. Data federation in the Biomedical Informatics Research Network: tools for semantic annotation and query of distributed multiscale brain data. AMIA Annu Symp Proc. 2008;1220 [PubMed] [Google Scholar]
- 5.Ozyurt IB, Keator DB, Wei D, Fennema-Notestine C, Pease KR, Bockholt J, Grethe JS. Federated web-accessible clinical data management within an extensible neuroimaging database. Neuroinformatics. 2010;8:231–249. doi: 10.1007/s12021-010-9078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott A, Courtney W, Wood D, de la Garza R, Lane S, King M, Wang R, Roberts J, Turner JA, Calhoun VD. COINS: An Innovative Informatics and Neuroimaging Tool Suite Built for Large Heterogeneous Datasets. Front Neuroinform. 2011;5:33. doi: 10.3389/fninf.2011.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King MD, Wood D, Miller B, Kelly R, Landis D, Courtney W, Wang R, Turner JA, Calhoun VD. Automated collection of imaging and phenotypic data to centralized and distributed data repositories. Front Neuroinform. 2014;8:60. doi: 10.3389/fninf.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus DS, Harwell J, Olsen T, Hodge M, Glasser MF, Prior F, Jenkinson M, Laumann T, Curtiss SW, Van Essen DC. Informatics and data mining tools and strategies for the human connectome project. Front Neuroinform. 2011;5:4. doi: 10.3389/fninf.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Kogan A, Cobia D, Alpert K, Kolasny A, Miller MI, Marcus D. Northwestern University Schizophrenia Data and Software Tool (NUSDAST) Front Neuroinform. 2013;7:25. doi: 10.3389/fninf.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Alpert K, Calhoun VD, Keator DB, King MD, Kogan A, Landis D, Talllis M, Potkin SG, Turner JA, Ambite JL. SchizConnect: Mediating Schizophrenia Neuroimaging Databases for Large-Scale Integration. NeuroImage. doi: 10.1016/j.neuroimage.2015.06.065. (Manuscript under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, Pacheco J, Albert M, Killiany R, Blacker D, Maguire P, Rosas D, Makris N, Gollub R, Dale A, Dickerson BC, Fischl B. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. NeuroImage. 2009;46:177–192. doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 14.Glover GH, Mueller BA, Turner JA, van Erp TG, Liu TT, Greve DN, Voyvodic JT, Rasmussen J, Brown GG, Keator DB, Calhoun VD, Lee HJ, Ford JM, Mathalon DH, Diaz M, O'Leary DS, Gadde S, Preda A, Lim KO, Wible CG, Stern HS, Belger A, McCarthy G, Ozyurt B, Potkin SG. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. Journal of magnetic resonance imaging : JMRI. 2012;36:39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson SD, Martone ME. Ontologies for Neuroscience: What are they and What are they Good for? Frontiers in neuroscience. 2009;3:60–67. doi: 10.3389/neuro.01.007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bug WJ, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird AR, Larson SD, Rubin D, Shepherd GM, Turner JA, Martone ME. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. 2008;6:175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larson SD, Martone ME. NeuroLex.org: an online framework for neuroscience knowledge. Front Neuroinform. 2013;7:18. doi: 10.3389/fninf.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner JA, Laird AR. The cognitive paradigm ontology: design and application. Neuroinformatics. 2012;10:57–66. doi: 10.1007/s12021-011-9126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poldrack RA, Kittur A, Kalar D, Miller E, Seppa C, Gil Y, Parker DS, Sabb FW, Bilder RM. The cognitive atlas: toward a knowledge foundation for cognitive neuroscience. Front Neuroinform. 2011;5:17. doi: 10.3389/fninf.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whetzel PL, Team N. NCBO Technology: Powering semantically aware applications. Journal of biomedical semantics. 2013;4(Suppl 1):S8. doi: 10.1186/2041-1480-4-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mejino JL, Rubin DL, Brinkley JF. FMA-RadLex: An application ontology of radiological anatomy derived from the foundational model of anatomy reference ontology. AMIA Annu Symp Proc. 2008:465–469. [PMC free article] [PubMed] [Google Scholar]
- 22.Rubin DL. Creating and curating a terminology for radiology: ontology modeling and analysis. Journal of digital imaging. 2008;21:355–362. doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckler AJ, Liu TT, Savig E, Suzek BE, Rubin DL, Paik D. Quantitative imaging biomarker ontology (QIBO) for knowledge representation of biomedical imaging biomarkers. Journal of digital imaging. 2013;26:630–641. doi: 10.1007/s10278-013-9599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood D, King M, Landis D, Courtney W, Wang R, Kelly R, Turner JA, Calhoun VD. Harnessing modern web application technology to create intuitive and efficient data visualization and sharing tools. Front Neuroinform. 2014;8:71. doi: 10.3389/fninf.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P, Ruttenberg A, Sansone SA, Soldatova LN, Stoeckert CJ, Jr, Turner JA, Zheng J consortium, O.B.I. Modeling biomedical experimental processes with OBI. Journal of biomedical semantics. 2010;1(Suppl 1):S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong YM, Dahlke C, Xiang Q, Qian Y, Karp D, Scheuermann RH. Toward an ontology-based framework for clinical research databases. Journal of biomedical informatics. 2011;44:48–58. doi: 10.1016/j.jbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poline JB, Breeze JL, Ghosh S, Gorgolewski K, Halchenko YO, Hanke M, Haselgrove C, Helmer KG, Keator DB, Marcus DS, Poldrack RA, Schwartz Y, Ashburner J, Kennedy DN. Data sharing in neuroimaging research. Front Neuroinform. 2012;6:9. doi: 10.3389/fninf.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breeze JL, Poline JB, Kennedy DN. Data sharing and publishing in the field of neuroimaging. Gigascience. 2012;1:9. doi: 10.1186/2047-217X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mejino JV, Jr, Agoncillo AV, Rickard KL, Rosse C. Representing complexity in part-whole relationships within the Foundational Model of Anatomy. AMIA Annu Symp Proc. 2003:450–454. [PMC free article] [PubMed] [Google Scholar]
- 30.Golbreich C, Grosjean J, Darmoni SJ. The Foundational Model of Anatomy in OWL 2 and its use. Artificial intelligence in medicine. 2013;57:119–132. doi: 10.1016/j.artmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Nichols BN, Mejino JL, Detwiler LT, Nilsen TT, Martone ME, Turner JA, Rubin DL, Brinkley JF. Neuroanatomical domain of the foundational model of anatomy ontology. Journal of biomedical semantics. 2014;5:1. doi: 10.1186/2041-1480-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sim I, Carini S, Tu SW, Detwiler LT, Brinkley J, Mollah SA, Burke K, Lehmann HP, Chakraborty S, Wittkowski KM, Pollock BH, Johnson TM, Huser V Human Studies Database, P. Ontology-based federated data access to human studies information. AMIA Annu Symp Proc. 2012;2012:856–865. [PMC free article] [PubMed] [Google Scholar]