Fig. 3.
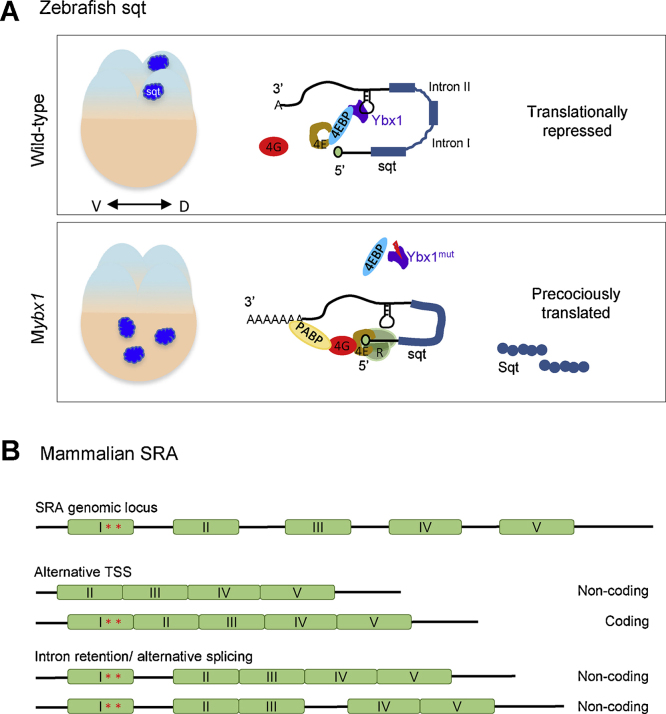
RNA processing facilitates dual function of cncRNAs. (A) Regulated splicing, polyadenylation, and translation in temporal partitioning of non-coding versus coding functions of sqt RNA in zebrafish. A 4-cell stage zebrafish embryo is depicted with dorsal progenitor cells (D) at the right side. In wild-type embryos, by the 4-cell stage, sqt transcripts are actively localized to 1 or 2 cells. In the schematic representation of sqt RNA, black lines represent UTRs, blue boxes represent the 3 coding exons and the blue lines represent the introns. Maternal RNA is not completely spliced, lacks a poly A tail and is translationally repressed by Ybx1. Ybx1 sequesters eIF4E (4E) either directly or in a complex with an eIF4E binding protein (4EBP) to prevent formation of the eIF4 translation pre-initiation complex and recruitment of ribosomes (R). In maternal ybx1 mutant (Mybx1) embryos, sqt RNA fails to localize and forms aggregates in the yolk. Maternal sqt RNA is precociously spliced, polyadenylated, and Sqt protein is translated prematurely in Mybx1 mutant embryos. This leads to premature activation of the Nodal/Squint pathway in Mybx1 mutants. (B) Alternative transcription start sites, intron retention and alternative splicing result coding and non-coding isoforms of SRA RNA. The SRA genomic locus consists of five coding exons, and exon I has two in-frame start codons (red asterisks). There is an alternative transcriptional start site (TSS) in exon I which leads to the production of a non-coding isoform of SRA. Alternative splicing leads to retention of intron I and sometimes intron III, and can also produce non-coding SRA isoforms. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)