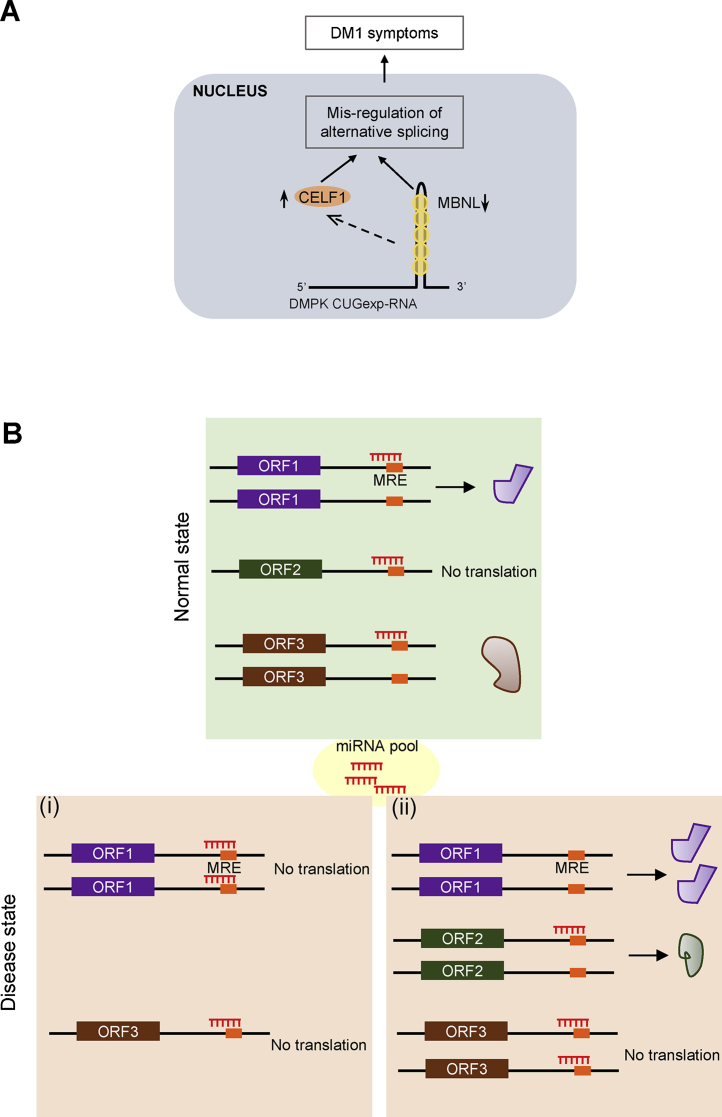
Fig. 4.
cncRNAs in disease. (A) Sequestration and modulation of regulatory proteins by DMPK mutant RNA in type I myotonic dystrophy. Variable length polymorphism resulting from increased number of CUG repeats in 3′UTR of DMPK gene leads to transcription of a toxic form of RNA (DMPK CUGexp-RNA). The CUG repeats form a stable stem loop (the hairpin in the DMPK CUGexp-RNA). The splicing factor MNBL (yellow) directly binds to the CUG repeats. Sequestration of MBNL makes it inactive. DMPK CUG-exp RNA stabilizes another splicing factor, CELF1 by indirect hyperphosphorylation. This leads to mis-regulation of alternative splicing and manifestation of type I myotonic dystrophy (DMI) symptoms. (B) ceRNAs as miRNA sponge. A group of mRNAs sharing a particular miRNA response element (MRE) function as ceRNAs and influence each other's translation. In a normal state, a limited pool of miRNAs can regulate the translation of a number of mRNAs in a ceRNA network. When the expression of one mRNA is changed, the redistribution of available miRNA molecules will result in a change in translational output of other mRNAs in the network, potentially leading to disease states. In this schematic, ORF2 functions as a miRNA sponge to regulate the translational output of ORF1 and ORF3. When ORF2 expression is reduced (disease state i) miRNA that was bound to ORF2 will be available to target other RNAs leading to repression of ORF1 and ORF3 while when ORF2 is overexpressed (disease state ii), translational repression of ORF1 and ORF3 by miRNA will be alleviated due to presence of more MREs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)