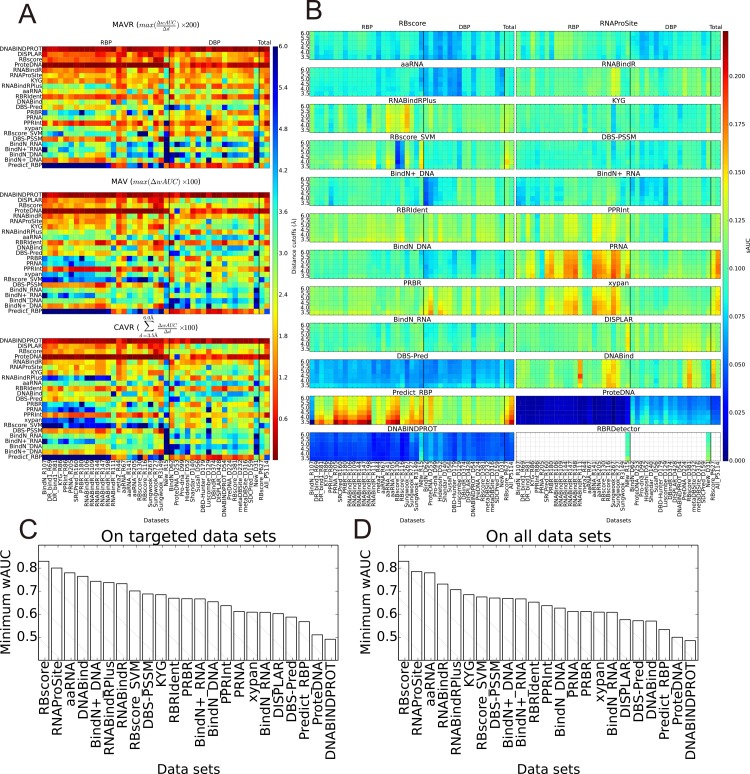
Fig 3. Accuracy variations resulted from binding site definition and data set bias.
A) Accuracy variation resulted from distance cutoff based definition of binding sites. Three metrics, MAVR, MAV and CAVR (see methods for details), were used to describe the distance cutoff resulted accuracy variation. The higher the values are, the less stable the predictor is, indicating a less reliable predictor; B) Standard deviation of AUC (sAUC) on all data sets. Similarly, the higher values of sAUC indicate an unstable predictor that cannot guarantee stable accuracy or it is more likely to have data set bias; C) Minimum wAUC of all programs on their targeted data sets: DNA binding site prediction programs are tested on DBP and minimum wAUC are plotted, while the same for RNA binding site prediction programs on RBP. As prediction accuracy of a predictor can vary with data set of assessment and distance cutoff to define binding sites, minimum wAUC demonstrates the bottom line of accuracy that a predictor can guarantee. D) Minimum wAUC of all programs on all data sets.