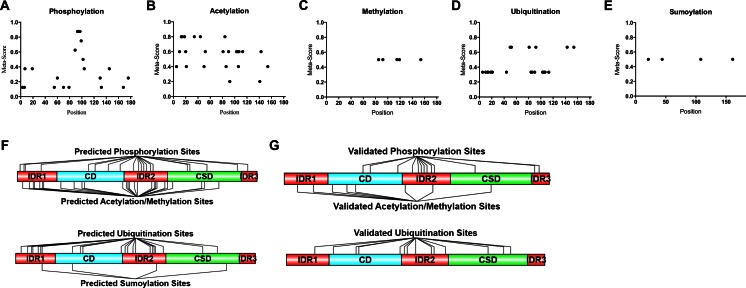
Fig. 8.
Characterization of IDR2 as the signal integration center through prediction of post-translational modifications. Prediction of post-translational modification sites on HP1γ was performed by compiling and statistically scoring linear motifs for phosphorylation (a), acetylation (b), methylation (c), ubiquitination (d), and sumoylation (e) as predicted by 20 different software programs. For each program, we considered sites for which the prediction score was above the cut-off that had been derived using a training set of modified sequences that have been experimentally validated. Subsequently, we developed a meta-prediction score by assigning a maximum score of 1 to sites that were predicted by all of the programs cited. Scores for other programs were numerically expressed relative to this maximum score of 1. This analysis revealed that predicted phosphorylation sites have high specificity potential near IDR2 of HP1γ. f Graphical representation of predicted post-translational modification sites. Results of this analysis revealed that post-translational modifications have the propensity to occur throughout the entire HP1γ sequence, but appear to be heavily localized to the IDR regions. g Graphical representation of experimentally validated post-translational modification sites listed on PhosphositePlus [57] and PHOSIDA [58]