Figure 1.
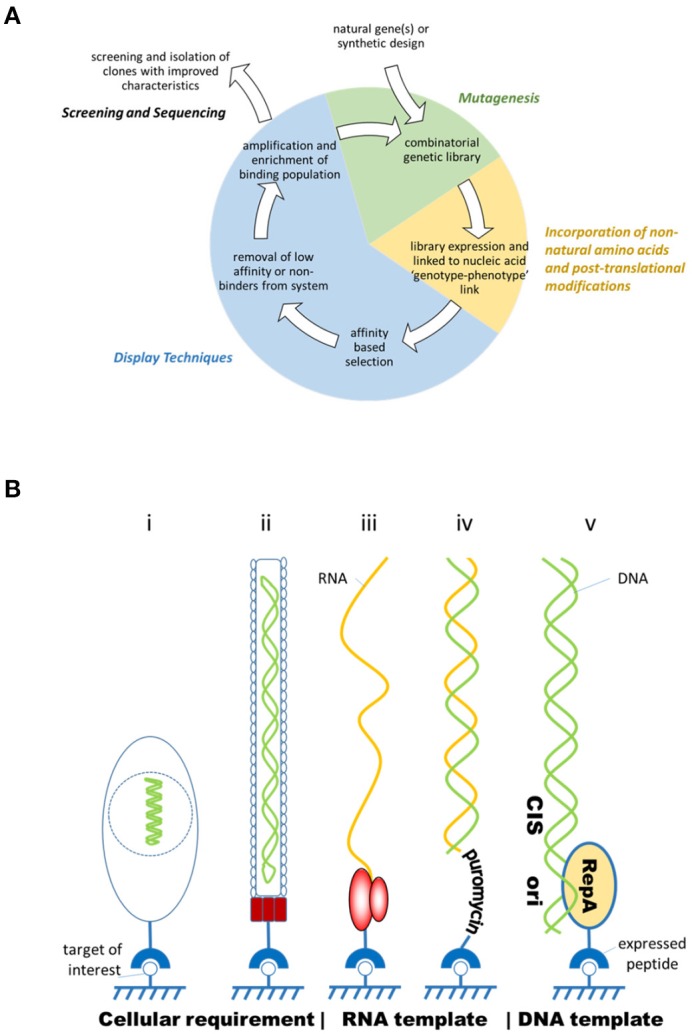
(A) Overview of the process of directed evolution. Combinatorial genetic libraries are created through mutagenesis, then transcribed and translated into peptides which retain a physical link with their encoding genetic information. Application of selection pressure, such as cycles of washing, amplification and further selection, enrich the binding clones within the population. Ultimately, binding peptides are identified through sequencing of the genetic material. (B) Diagram showing different formats of display where the peptide is expressed on the surface of a yeast or bacterium (i) (yeast/bacterial display); or a bacteriophage (ii) (phage display); or in complex with a ribosome (iii) (ribosome display); linked to RNA through puromycin (iv) (mRNA display); or bound to DNA through the cis activity of a DNA binding protein (v) (CIS display). In all these examples the peptide is associated with its coding sequence maintaining a genotype-phenotype link and is free to associate to its target (adapted from Ullman et al., 2011).
